ISSN: 0973-7510
E-ISSN: 2581-690X
Foot-and-mouth disease virus (FMDV) is small RNA virus from Picornaviridae family; genus Aphthovirus. FMDV causes maximum levels of infectivity in cattle and harmful socioeconomic effects. The present report attempted to design vaccine candidate from the polyprotein of FMDV to stimulate protective immune response. The IEDB server was used to predict B and T cells epitopes that were linked via GPGPG and YAA linkers, respectively. Mycobacterium tuberculosis 50S ribosomal protein was exploited as an adjuvant and a six histidine-tag sequence was linked to the carboxyl end of the vaccine for purification and identification. The predicted vaccine comprised 313aa and was antigenic and not allergic. Moreover, the vaccine was acidic and showed stability and hydrophilicity. Vaccine secondary and tertiary structures were predicted. The tertiary structure was refined to ameliorate the quality of the global and local structures of the vaccine. Vaccine model validation was performed and the final quality score of the structural model was computed. The validated model was used for molecular docking with bovine (N*01801-BoLA-A11) allele. Docking process in terms of binding free energy score was significant. Vaccine solubility was investigated based on the protein of E. coli and the stability was based on the disulfide bonding to lessen the entropic and mobile points in vaccine. Lastly, the in silico cloning ensured the proper cloning and best translation of the DNA of vaccine in molecular vectors.
FMDV, FMD, B lymphocyte Cells, T lymphocyte Cells, Peptide Vaccine
Foot-and-mouth disease virus (FMDV) is an RNA-virus (single-stranded RNA), non-enveloped, icosahedral shape with a smooth soft surface, about 30nm diameter and approximately 8.5 kb nucleotides long. FMDV was the initial realized viral in the genus Aphthovirus that related to Picornaviridae family.1 The name Picornaviridae (indicated small RNA) referred to the small size of the virus genomic matter, while the genus aphtho (from the Greek aphtha) refers to the vesicles in the mouth of infected animals.2,3
FMDV cause Foot-and-mouth disease (FMD) which is a highly contagious disease of footed cloven mammals worldwide like cattle, water buffalo, goats, sheep, pigs, deer, and bison.4-6 The disease has maximum levels of infectivity in all animals and demonstrated harmful social and economic impacts in affected regions.5 The transmission of FMD is possible by direct/ indirect adherence with diseased and/ or contaminated fomites of the animals. The virus can spread by multiple routes such as aerosolized virus inhalation, contaminated feed and water, and through skin wounds and abrasions or orifices mucous membranes. Sexual transmission was observed in African buffalo by SAT type viruses.6,7 Vaccinated animals or animals with natural immunity can become carriers of the virus for several months or less, however, some animals became permanently carriers of the infection for several years.6 The incubation time is 2 to 3 days up to two weeks, the infected animals might spread the infection before providing the symptoms of the disease.1 The disease demonstrated signs such as high fever, vesicles formation in and around buccal cavity, teats and round feet that pop and turn into red areas called erosions.8-10 Pain and discomfort from the vesicles and erosions lead to other symptoms such as depression, loss of appetite, excessive salivation, lameness, aversion to move or stand and reduction in milk production.11 The severity of clinical signs depends on the virus strain, exposure dose, age and strain of affected animals. Mortality is most seen in young than adult animals from a multifocal myocarditis, so called tiger heart.6 Human infections with FMDV rarely occurred and only about 40 cases were reported since 1921.12
FMD is considered as an endemic disease in most of African, South American and Asian lands. The disease demonstrated amazing capability to cross the international borders resulting in epidemics in other free zones areas.9,13 FMD outbreak resulted in considerable costs, particularly the cost of control, hindering animal movements, prevention measures and prohibition of exportation.3 The major economic effect in endemic countries is reduction in milk production and livestock growth due to infertility caused by the virus, mortality in young animals and abortion.14,15 The impact of FMD outbreaks were obvious in Sudan, where the disease occurred during the year 2002 in dairy farms of Khartoum state. The estimated loss was in the high mortality rate among the milking cows and calves with vast prominent reduction in milk production as major factor of the disease cost.15
The FMDV comprises 7 serotypes (SAT1, SAT2, SAT3, O, A, C and Asia1) in addition to multiple subtypes occur during virus evolution.4,5 in Africa, the most prevalent serotypes were A, O, SAT1 and SAT2.2 The first reported FMD cases in Sudan were in the year 1903. Since then, the outbreaks were reported annually in Sudan particularly during winter season.2 For instance, the serotype O was initially isolated, followed by SAT1, serotype A and lastly serotype SAT2.16
FMDV belongs to the RNA single stranded, positive sense viruses with an 8.5 kb. The virus genome has three main regions: (a) 5′ regulatory noncoding region (b) coding region of the polyprotein (comprises regions called L, P1, P2, and P3) (c) 3′ regulatory noncoding region.17 The polyprotein mainly translated from the viral genome as a single protein and then post translationally modified by cleavage via viral proteases resulting in four structural proteins named VP1, 2, 3 and 4. These four proteins are essential for viral assembly, provision immunogenicity to the virus and are significant for binding of the virus to host cell receptors.18-20 Other ten mature nonstructural proteins are essential for the proteolytic activities, virus replication, fitness and virulence.9,17,19,21 FMDV polyprotein processing is mediated by L, 3C, and 2A.19 The capsid consists of sixty copies from each structural protein (VP1, 2, 3 and 4).22 The VP4 is considered an internal protein while the others located on the surface of the virion. VP1 is the immunogenic structural protein within the polyprotein carrying the neutralizing antigenic sites because its GH loop sticked out to surface forming large part of virus surface. Among FMDV serotypes VP2 and VP3 are quite conserved while VP4 is most conserved one.1,5,17,22
Previous reports concerning the antigenicity and immunogenicity of FMDV guaranteed the significance of VP1 as important antigenic determinants on the virion surface. The antigenic diversity of FMDV was augmented by time, and this attributed to immunological pressures on the virus.10 The enhanced action of GH loop combined with the action of C vicinity of VP1 protein provided multiple antigenic sites (1, 2 and 3 and 4) with important amino acids residues at multiple positions that enhanced the immunogenicity of the virus. Previous studies showed that antigenic site 1 as an immunodominant and obvious target region for synthesis of peptide vaccines due to its structural linearity.10,22-24
The present study attempted to design peptide vaccine candidate against FMDV exploiting the polyprotein as an immunogen enhancing immune response and helping in production of peptide vaccine with powerful immune protection.
Retrieval of Viral Polyproteins Sequences
A total of 14 FMDV strains viral polyproteins were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/protein/?term=Footandmouth+disease+ virus++type+O). Fourteen polyprotein sequences retrieved from Southern Asia most of them from China (Table 1).
Table (1):
Retrieved strains and their accession numbers and area of collection.
Accession Number |
Date of collection |
Country |
|---|---|---|
*NP_658990.1 |
1997 |
Taiwan |
APH81273.1 |
2013 |
China |
APH81272.1 |
2013 |
China |
ADV52246.1 |
2005 |
Malaysia |
ADV52244.1 |
2001 |
Malaysia |
ADU15820.1 |
2000 |
China |
AAT01778.1 |
1997 |
Taiwan |
AAT01776.1 |
1999 |
Taiwan |
ABM63320.1 |
2007 |
China |
ABB69024.1 |
2005 |
China |
AAG45408.1 |
1997 |
Taiwan |
ABY91241.1 |
2001 |
China |
AAT97073.1 |
2001 |
China |
AAM91949.2 |
2002 |
China |
*Ref sequence
Strains Ancestral Analysis
Ancestral analysis of the strains sequences was performed to determine the relatedness and the common ancestral origin of the strains using tools at (http://www.ebi.ac.uk/Tools/msa/clustalo/ ).
Analysis of Strains Conservancy
The conserved regions or the conserved sequences between the strains was analyzed using the alignment features in the Clustal-W in the BioEdit software (version 7.0.9.1).25
B-cell Epitope Prediction
For the detection of candidate epitopes from FMDV polyprotein, various prediction methods using Immune Epitope Database (IEDB) resource at (http://www.iedb.org/)26,27 were used. Epitopes interacting with B cells are parts of an immunogene that interacting against B lymphocytes. Thus the B-lymphocytes proliferated and differentiated forming memory antibody-secreting plasma cells. Thus such cells are being antigenic and accessible.28 The reference sequence of FMDV polyprotein (NP_658990 with 2322aa) was submitted to Bepipred, Emini surface accessibility and Kolaskar antigenicity tools in IEDB for predicting B cells epitopes.
Bepipred Prediction Tool
BepiPred tool in the IEDB server at (http://toolsiedb.ofg/bcell/ ) was used to analyze linear epitopes from the input sequence of the polyprotein reference sequence.26
Emini Prediction Tool
By using Emini surface accessibility prediction tool in IEDB server at (http://tools.immuneepitope.org/tools/bcell/iedb), only the linear epitopes located on the surface of polyprotein reference sequence were predicted.26
Kolaskar and Tongaonkar Prediction Tool
The Kolaskar and Tongaonkar antigenicity method in the IEDB server at (http://tools.immuneepitope.org/bcell/) was used to assess the antigenic epitopes among the linear and surface accessible epitopes.26
Prediction of Epitopes Interacting with MHC Class I
In this study the interacting epitopes against MHC class I were only predicted as the IEDB server not yet assembled MHC class II data for bovine spp. Thus analysis of MHCI interacting epitopes was investigated using the tool at (http://tools.iedb.org/mhci/). Artificial Neural Network (ANN) was used as prediction tool with epitopes length of nine amino acids.26,28 Epitopes-alleles scoring threshold was ≤ 500 half-maximal inhibitory concentration (IC50).29
Assessing Antigenic, Nonallergenic and Nontoxic Epitopes
Vaxijen server at (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html ) was used to analyze the antigenic epitopes of the B and T cells with the default threshold (0.4). Allertop30 and toxinpred31 servers were exploited to determine the nonallergenic and nontoxic epitopes, respectively.
Vaccine Assemblage
The structure of the vaccine construct was made by combination of the predicted epitopes of B and T cytotoxic cells. The immunogenicity of the vaccine was enhanced by combining the 50S ribosomal protein L7/L12 of Mycobacterium tuberculosis (uniprot P9WHE3) as an adjuvant. This adjuvant was added at the amino terminal of the vaccine and separated form the vaccine sequence by EAAAK linker. The GPGPG and YAA linkers were used to separate and fuse the B cells and T cells epitopes, respectively. A set of six histidine molecules (6 his-tag sequence) was added on the carboxyl terminal of the vaccine for purification and identification.
Vaccine Physio-chemical Properties
To compute the physio-chemical properties of the vaccine, the Expasy ProtParam server (https://web.expasy.org/protparam/) was used. Multiple parameters were computed such as the vaccine molecular weight, atomic composition, theoretical isoelectric point, estimated half-life, aliphatic index, instability index, extinction coefficient and grand average of hydropathicity (GRAVY).32
Prediction of the Secondary Structure and Solvent Accessibility of the Vaccine
Vaccine secondary structure (SS), solvent accessibility (ACC) and disorder regions (DISO) in the vaccine sequence were assessed based on Raptor X server.33-34 The SS comprises the β-sheet (E), a-helix (H) and coiled regions (C) in the structure. The ACC were assessed using the solvent accessibility tool present in Raptor X server.35 The ACC results were demonstrated as E, M and B for exposed, medium and buried residues, respectively. While the cutoff value at 0.25 was used to assess the DISO regions.
Vaccine Tertiary Structure Prediction, Refinement and Validation
I-TASSER server was used to obtain the 3D vaccine structure in PDB format.36 The PDB structure was further refined by GalaxyRefine server to ameliorate the overall quality and validate the local and global structure of the vaccine.37 Model validation was performed through Saves Ramchandran plot (https://saves.mbi.ucla.edu/). The ProSA-web server38 was further used to compute the overall quality score of the input protein PDB structure.
Solubility and Stability of the Vaccine Candidate
Protein sol server for detection of proteins solubilities was used to predict the vaccine solubility in terms of scaled solubility value (QuerySol) in relation to the E. coli proteins population average dataset (PopAvrSol) of 0.45.39 Proteins that demonstrated solubility scores greater than E. coli experimental dataset (0.45) were considered as high soluble proteins and vice versa. For protein stability, the Disulfide by Design 2 (DbD2) is software used for designing disulfide bonds in protein via rapid assessing protein geometrical structure suitable for disulfide bonds formation, assuming the amino acids residues were converted or mutated into cysteines.40 This software was used to predict disulfide bonds that assisted in dynamics and interactions analysis of the vaccine. All potential parameters in the software were set to default upon prediction.
Molecular Docking of the Vaccine and Bovine MHC-I N*01801 (BoLA-A11)
The docking process was performed using the ClusPro 2.0 server. The server used docking automation and discrimination methods for prediction of proteins-receptors complexes.41 The PDB file of the predicted vaccine was docked with the bovine MHC-I N*01801 (BoLA-A11) allele as a receptor molecule (PDB: 3PWU) with the receptor chain A. Multiple docking complexes were obtained and the one with the best binding energy was chosen and visualized using PyMOL software (www.pymol.org).
In silico Cloning
The vaccine protein sequence was first reversed to DNA sequence using JCAT server (Java Codon Adaptation Tool) at (http://www.prodoric.de/JCat). The best codons score in the DNA sequence between 1.0 but more than 0.8 is considered as a favorable score with GC percentage between 30% to 70%.42 The sequences of BamH1 and Xho1 restriction enzymes were added to the vicinities of the DNA molecule. The SnapGene cloning software was used to clone the DNA into pET28a (+) cloning vector.
Ancestral Analysis and Epitopes Conservancy
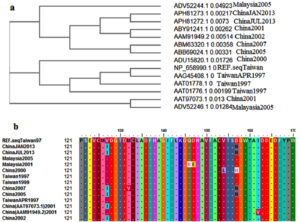
As shown in Figure 1a, the ancestral analysis showed closed relationship between the retrieved strains. The Malaysia strains were closely related to the Chines and Taiwan strains. However despite the closed relationship between the retrieved strains molecular divergence was observed between the strains. MSA of the retrieved strains was represented in Figure 1b and predicted epitopes showed conservancy within the aligned sequences. Regions showed conservancy demonstrated identical amino acid sequences between retrieved strains sequences.
Figure 1. (a) showed the ancestral analysis of the retrieved strains. (b) Showed MSA between the strains. Dots indicated conservancy while letters within the sequences indicated mutated regions
B-cells Epitopes Prediction
Bepipred Prediction Tool
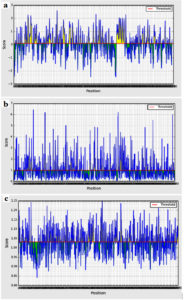
Bepipred binding average score of the predicted epitopes to B cells was between 2.597 and -2.476 as maximum and minimum scores, respectively. Sum of 52 epitopes were shown as linear epitopes eliciting B lymphocyte with the threshold value of 0.350 (Figure 2a). However, only 24 epitopes were shown to be linear conserved epitopes.
Figure 2. B cell epitopes prediction. (a) Bepipred linear tools, (b) Emini prediction tool for surface accessibility (c) Kolaskar and Tongaonkar tool for antigenicity prediction. The red line indicated the default threshold for each tool
Emini Prediction Tool
The surface accessibility average score was 4.134 (ranging between 7.795 and 0.035 as maximum and minimum, respectively). Only 17 out of the 24 linear conserved epitopes were potentially surface accessible passing the default threshold 1.000 (Figure 2b).
Kolaskar and Tongaonkar Tool
The antigenicity average score was 1.031 (ranging between 1.250 and 0.842 as maximum and minimum, respectively). Only 9 epitopes out of the 17 surface accessible epitopes passed the threshold of 1.031 (Figure 2c) and were considered as antigenic epitopes.
Taken together, only nine epitopes were predicted as B cells epitopes as they demonstrated linearity, surface accessibility and antigenicity. Also, these epitopes were antigenic in vaxigen web server and nonallergic and nontoxic in allertop and toxinpred servers, respectively. The nine epitopes and their features were shown in Table 2.
Table (2):
The predicted B cell epitopes.
Peptide |
Start |
End |
Length |
Emini surface accessibility (1.000) |
Kolaskar antigenicity (1.031) |
Vaxigen antigenicity (0.4) |
Allergenicity |
Toxicity |
|---|---|---|---|---|---|---|---|---|
SRPSEV |
119 |
124 |
6 |
1.558 |
1.032 |
0.6640 |
non-allergen |
Non-Toxic |
LVTTDPKTADPVYGKVFNPPRNL |
518 |
540 |
23 |
1.407 |
1.036 |
0.4144 |
non-allergen |
Non-Toxic |
AADYAYT |
668 |
674 |
7 |
1.063 |
1.041 |
1.0603 |
non-allergen |
Non-Toxic |
ESADPV |
730 |
735 |
6 |
1.001 |
1.04 |
0.9734 |
non-allergen |
Non-Toxic |
PTAYHK |
828 |
833 |
6 |
2.142 |
1.039 |
1.4858 |
non-allergen |
Non-Toxic |
APAPSK |
1200 |
1205 |
6 |
1.457 |
1.033 |
0.4999 |
non-allergen |
Non-Toxic |
TDSVWYCPPDPDHFD |
1242 |
1256 |
15 |
1.088 |
1.041 |
0.4067 |
non-allergen |
Non-Toxic |
GPYAGPLERQKPLKVKA |
1569 |
1585 |
17 |
2.15 |
1.038 |
0.5034 |
non-allergen |
Non-Toxic |
HIDPEPHH |
1844 |
1851 |
9 |
1.67 |
1.039 |
2.0013 |
non-allergen |
Non-Toxic |
MHC Class I Interacting Epitopes Prediction
A total of 17 epitopes were shown interacting with various MHCI alleles based on IC50≤500 and ANN- align method. Among these seventeen epitopes only four epitopes passed the criteria of being antigenic nontoxic and nonallergic epitopes (Table 3).
Table (3):
The predicted cytotoxic T cell epitopes.
Peptide |
Start |
End |
Interacted alleles |
IC50 |
Vaxigen antigenicity (threshold (0.4) |
Allergenicity |
Toxicity |
|---|---|---|---|---|---|---|---|
AARSKDPVL |
1030 |
1038 |
BoLA-HD6 |
11 |
1.1611 antigenic |
non-allergen |
Nontoxic |
DHKGVYGSL |
372 |
380 |
BoLA-T2C |
227.92 |
0.5795 antigenic |
non-allergen |
Nontoxic |
GQHEAAIEF |
1442 |
1450 |
BoLA-D18.4 |
43.91 |
0.8012 antigenic |
non-allergen |
Nontoxic |
TQYSGTINL |
603 |
611 |
BoLA-D18.4 |
32.65 |
0.4008 antigenic |
non-allergen |
Nontoxic |
BoLA-HD6 |
14.78 |
Structure of the Assembled Vaccine
The B and T cell epitopes used to construct the vaccine were nine the linear B-cell epitopes and the four T cytotoxic cell epitopes from FMDV polyprotein. The final vaccine sequence comprises 313aa and was shown to be antigenic (0.4999) and nonallergen (Table 4).
Table (4):
The physical and chemical properties, antigenicity and the number of the predicted transmembrane helices of the vaccine protein.
Vaccine Sequence* |
Molecular weight (D) |
Instability index# |
Aliphatic index |
Theoretical PI |
No amino acids |
Extinction Coefficient |
GRAVY† |
Vaxijen antigenicity$ |
TMHs |
MAKLSTDELLDAFKEMTLLELSDF KKFEETFEVTAAAPVAVAAAGA APAGAAVEAAEEQSEFDVILEAAG DKKIGVIKVVREIVSGLGLKEAKD LVDGAPKPLLEKVAKEAADEAKA KLEAAGATVTVKEAAAKAARSKD PVLGPGPGDHKGVYGSLGPGPGG QHEAAIEFGPGPGTQYSGTINLYAA SRPSEVYAALVTTDPKTADPVYGK VFNPPRNLYAAAADYAYTYAAES ADPVYAAPTAYHKYAAAPAPSK YAATDSVWYCPPDPDHFDYAAGP YAGPLERQKPLKVKAYAAHIDPEP HHHHHHHH |
32791.81 |
34.99 |
75.08 |
5.44 |
313 |
30830 |
-0.279 |
0.4999 |
0 |
*In the vaccine sequence, the linkers were shown in bold; #instability index <40 considered the protein stable; †GRAVY negative sign indicated the protein is hydrophilic; $the threshold for the Vaxijen antigenicity is 0.4; THMs: Transmembrane helices.
Vaccine Physio-chemical Properties
ProtParam server was used to compute multiple physio-chemical properties of the vaccine. The results of these features are included in Table 4. In brief, the vaccine was shown to be acidic and hydrophilic in nature. Moreover, the instability index was less than 40, thus classifies the vaccine as stable.
Secondary Structure and Solvent Accessibility Prediction
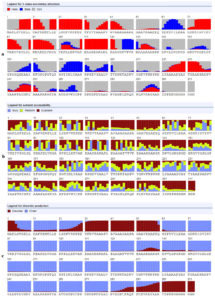
Figure 3 provided the detailed prediction of SS3, ACC and the DISO. The SS3 provided 29%, 10%, and 60% of the residues as alpha, beta and coiled structures, respectively. The ACC showed 55%, 23% and 20% of the residues as E, M and B residues, respectively. Sums of 56 residues (17%) were predicted as DISO regions.
Figure 3. a- secondary structure prediction, b- solvent accessibility prediction and c- ordered and disordered residues of the vaccine construct
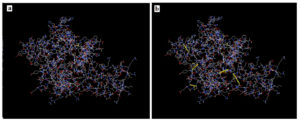
Prediction of Tertiary Structure (3D), Refinement and Validation
Figure 4a showed the 3D of the vaccine in I-TASSER sever while Figure 4b provided the refined structure by Galaxyrefiner server. Structural refinement was assessed to meliorate the structure quality. The Z-score of proSA web program was -3.54 representing model fine quality (Figure 4c). Ramachandran plot assessed the stability of the vaccine post-refinement. The favored region in the plot comprises 91.9% of the residues; the allowed region comprises 3.1% of the residues with only 5.0% of the residues in the disallowed region (Figure 4d).
Figure 4. (a) the 3D tertiary structure of the vaccine, (b) the vaccine structure after refinement. (c) ProSA-web Z-score was −3.54 and (d) validation of vaccine (91.9% of the residues in favored region, 3.1% in the allowed region and 5.0% in the outlier region)
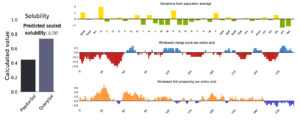
Solubility and Stability of the Vaccine
Figure 5 provided solubility QuerySol of 0.737 in comparison to the PopAvrSol (0.45). In this regard, solubility of the vaccine protein was higher than that E. coli (0.45), indicating solubility of the vaccine. For the stability total of 46 pairs of amino acid residue were implicated in disulfide engineering. Figure 6 provided the most 5 regions that were considered as high mobile regions in the protein mutated to build cys-cys disulfide bonds based on the chi3 residues and energy value lower than 2.0. The five residue pairs were 63PHE-113ALA; 131GLU-210VAL; 233ALA-236SER; 239PRO-241TYR and 280GLY287GLU.
Figure 6. Disulfide bond engineering in the vaccine protein. (a) is the original form and (b) the mutant form. Five disulfide bond regions were shown in golden sticky forms in the mutant form
Molecular Docking
Cluspro server validated docking process depending on three computational steps. Firstly: scanning billions of conformations between the receptor and ligand to obtain a rigid body docking molecules. Secondly: RMSD-based clustering of 1000 lowest energy structures to obtain high clusters representing the most probable model of the complex. Thirdly: elimination of energy used in space collisions minimizing the docking of the ligand (vaccine) to receptor. Figure 7 showed biologically significant results of the docking as indicated by free binding energy score of -1037.6. The negatively binding energy score value demonstrated the strong binding between bovine MHC-I N*01801 (BoLA-A11) allele and vaccine protein.
Figure 7. molecular docking of the vaccine protein (cyan color) against N*01801 (BoLA-A11) allele (yellow color). Region of the interaction was shown and zoomed in boxes and the bonding interaction (magenta color) was shown in the zoomed box
In silico Molecular Cloning
The in silico cloning provided CAI-value of 1.0, showing improved in the codon adapted sequence and the GC-content was 54.05%. Figure 8), showed the cloning of the DNA into pET28a (+) vector between BamH1 and Xho1restriction enzymes.
Vaccines formulated against FMDV were considered as the first animal vaccines initiated, as inactivated whole-virus was used as conventional immunization method. For instance, inactivated vesicular fluids treated with formaldehyde derived from the tongue of FMD infected cattle was developed as a vaccine.42 However the variability in the disease regions hindered the development of the vaccines that provoke protection against the virus serotypes.9 Currently vaccine product that is still in use was developed by inactivation FMDV antigen in oil-adjuvants in the 1970s.9,42 Commonly vaccine products exposed to quality control investigation to guarantee identity, safety, sterility, efficacy, potency and purity.43 Thus creation of such vaccine requires a massive degree of safety to avoid virus spreading from infected animals. Moreover, the inactivated vaccines against the FMDV failed to differentiate between vaccinated and infected animals.43,44 In addition to that, FMD conventional vaccines provided better immunity against clinical infection but failed to induce wide term of protection. Also, these vaccines require to repeat the vaccination process and require the inclusion of the new virus strains and serotypes in vaccine formulation to guarantee ideal immunity levels.45
FMD polyprotein was determined as best antigenic locus in FMDV genome, leading to peptides vaccines design as an alternatives, and can reach 95% purity. For instance purified polyproteins obtained from twelve FMD viruses and cloned in E. coli provided protection against FMD infection in both cattle and swine. The predicted peptides from VP1 were demonstrating generation of neutralizing antibodies but only partial in cattles.43 Some scientific reports united these peptides with FMDV T-cells peptides and results showed enhanced protection in swine.43,46 Nevertheless, such peptides to control FMD requires further analysis and developmental evaluation.43
In this study, only epitopes with 100% conservancy from the virus polyprotein were analyzed and chosen as highly immunogenic epitopes against B and T cells. For B cell epitopes, the results demonstrated that the thresholds of the proposed epitopes were greater the thresholds provided by the prediction tools in IEDB. Also, and most importantly, the produced vaccine should provoke the CD8+T cell mediated immunity which is considered as long lived from the crossed serotypes.47 Therefore the reference polyprotein was also analyzed to predict T cytotoxic cell epitopes interacting with different MHC-I bovine antigen (BoLA)-alleles. The overall results of the prediction provided the precise election of the epitopes interacting against B and T lymphocytes. The predicted epitopes from B and T cells were used to build a vaccine with linkers and adjuvant from the virus polyprotein.48 The linkers and adjuvant were shown to enhance minimal junctional immunogenicity and augment the expression level and ameliorate the vaccine bioactivity.49-52 The proposed vaccine was antigenic with no allergenicity and toxicity with favorable chemical and physical properties.
The secondary and tertiary structures of the vaccine and their folding stability can directly influence the conformational epitopes and availability of B-cell epitopes.53 Thus for our chimeric vaccine the secondary and tertiary structures were analyzed for stability. The results demonstrated high integrity of the vaccine candidate by containing extended strands, alpha helices, beta turns rather than having any other unambiguous states. Moreover, the vaccine 3D structure amelioration on Ramachandran plot demonstrated desirable prediction characteristics, thus indicating satisfactory quality of the designed model.
The solubility of the vaccine was analyzed based on and in comparison to solubility of E. coli proteins.39 The predicted vaccine was shown to be soluble. For stability, the structural disulfide bonding were reported to decrease the possible conformational numbers for a given protein, resulting in reduced entropy and enhanced protein thermostability.54,55 The predicted vaccine showed five possible regions for disulfide bonding construction giving the stability of the vaccine protein.
The vaccine protein was docked against bovine allele protein to provide the favorable interaction between the two proteins. The strong binding (docking) between bovine MHC-I N*01801 (BoLA-A11) allele and the vaccine protein showed negative value of the docking process resulting in strong binding between bovine allele and the chimeric vaccine. Moreover, the vaccine was cloned in suitable vector for immunoreactivity.56 E. coli systems are preferable choice for molecular cloning and production of recombinant protein.57-58 The vaccine protein showed high-level expression and translation in E. coli.
FMDV has high prevalence in the world for long time, so developing an effective vaccine is a necessity. Multiple peptides that would be a powerful effective vaccine against FMDV from the polyprotein were proposed. We recommended formulating a peptide vaccines including other serotypes of the virus. This is because there would be a possibility for existing common conserved epitopes from different strains of the virus. An in vitro and in vivo analysis of the predicted peptides is required to demonstrate their efficacy.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RMSA and YAA conceived and designed the study. YAA and KAA performed data analysis and coordinated the study. RMSA and YAA wrote the manuscript. KAA edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The (retrieved strains from the NCBI, data analysis from the IEDB server) data used to support the findings of this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Kadabo AHI. Sero-prevalence and risk factors of foot and mouth disease virus (sero-type A) in cattle in Khartoum State-Sudan (Dissertations). Sudan University of Science and Technology. 2000. http://sustech.edu
- Habiela M, Ferris NP, Hutchings GH, et al. Molecular characterization of foot-and-mouth disease viruses collected from Sudan. Transbound Emerg Dis. 2010; 57(5):305-314.
Crossref - Kenubih A. Foot and Mouth Disease Vaccine Development and Challenges in Inducing Long-Lasting Immunity: Trends and Current Perspectives. Vet Med (Auckl). 2021;12:205-215.
Crossref - Arzt J, Baxt B, Grubman MJ, et al. The pathogenesis of foot-and-mouth disease ii: viral pathways in swine, small ruminants, and wildlife; myotropism, chronic syndromes, and molecular virus-host interactions. Transbound Emerg Dis. 2011;58(4):305-326.
Crossref - Wenming Liu BY, Mingxia W, Weifeng L, et al. Identification of a conserved conformational epitope in the VP2 protein of foot-and-mouth disease virus. Arch Virol . 2017;162(7):1877-1885.
Crossref - El Damaty HM, Fawzi EM, Neamat-Allah ANF, et al. Characterization of foot and mouth disease virus serotype SAT-2 in swamp water buffaloes (Bubalus bubalis) under the Egyptian smallholder production system. Animals (Basel). 2021;11(6):1697.
Crossref - OIE. OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals [Mammals, Birds and Bees). 7th ed. online: OIE – World Organisation for Animal Health. 2016.
- Namatovu A, Tjornehoj K, Belsham GJ, et al. Characterization of foot-and-mouth disease viruses (FMDVs) from Ugandan cattle outbreaks during 2012-2013: evidence for circulation of multiple serotypes. Plos One. 2015;10(2):e0114811.
Crossref - Noureldin AMMA. Prevalence and risk factors of foot and mouth disease of cattle in Khartoum State – Sudan. Sudan University of Science and Technology; 2014.http://sustech.edu/
- Ularamu HG, Ibu JO, Wood BA, et al. Characterization of foot-and-mouth disease viruses collected in Nigeria between 2007 and 2014: evidence for epidemiological links between west and East Africa. Transbound Emerg Dis. 2017;64(6):1867-1876.
Crossref - APHIS. Foot-and-Mouth Disease In: Agriculture USDo, editor.: Animal and Plant Health Inspection Service 2013:2.
- Fiebre Aftosa. Foot and Mouth Disease. OIE World Organisation for Animal Health. 2014. http://www.cfsph.iastate.edu/Factsheets/pdfs/foot_and_mouth_disease.pdf. 2014. Revised: March 2021
- Mahapatra M, Hamblin P, Paton DJ. Foot-and-mouth disease virus epitope dominance in the antibody response of vaccinated animals. J Gen Virol. 2012;93(Pt 3):488-493.
Crossref - Knight-Jonesa TJD, Rushtonb J. The economic impacts of foot and mouth disease – What are they, how big are they and where do they occur? Prev Vet Med. 2013;112(3-4):161-173.
Crossref - El-Hussein AM, Daboura A. Economic impact of an outbreak of Foot and Mouth Disease in Khartoum State, Sudan. Vet World. 2012;5(4):219-222.
Crossref - Habiela MMA, Alamin G, Raouf YA, Ali YH. Epizootiological study of foot and mouth disease in the Sudan: the situation after two decades. Veterinarski Arhiv. 2010;80(1).
- Longjam NDR, Sarmah AK, Tayo T, Awachat VB, Saxena VK. A brief review on diagnosis of foot-and-mouth disease of livestock: conventional to molecular tools. Veterinary Medicine International. 2011;2011:905768.
Crossref - Pandya M, Rasmussen M, Hansen A, et al. A modern approach for epitope prediction: identification of foot-and-mouth disease virus peptides binding bovine leukocyte antigen (BoLA) class I molecules. Immunogenetics. 2015;67(11-12):691-703.
Crossref - Carrillo C, Tulman ER, Delhon G, et al. Comparative genomics of foot-and-mouth disease virus. J Virol. 2005;79(10):6487-6504.
Crossref - Han SC, Hui CS, Shi QJ, et al. Productive entry of foot-and-mouth disease virus via macropinocytosis independent of phosphatidylinositol 3-kinase. Sci Rep. 2016;6(1):19294.
Crossref - Yao-Zhong DHC, Jie Z, Jian-Hua Z, et al. An overview of control strategy and diagnostic technology for foot and mouth disease in china. Virol J. 2013;10:78.
Crossref - Mahapatraa SM, Madhanmohanb M, Subramaniamc S, et al. Antigenic and genetic comparison of foot-and-mouth disease virus serotype O Indian vaccine strain, O/IND/R2/75 against currently circulating viruses. Vaccine Elsevier. 2015;33(5):693-700.
Crossref - Wang Y, Shen Q, Jiang Y, et al. Immunogenicity of foot-and-mouth disease virus structural polyprotein P1 expressed in transgenic rice. J Virol Methods. 2012;181(1):12-17.
Crossref - Hassan FR, Shaifur Rahman MD, Rahman KMT et al. In-Silico evaluation of the capsid proteins of FMDV as potential vaccine candidates. Computational Biology and Bioinformatics. 2015;3(1):6-20.
Crossref - Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947-2948.
Crossref - Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7(1):439.
Crossref - Hall TA, editor BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series; 1999: (London): Information Retrieval Ltd., c1979-c2000.
- Vita R, Overton JA, Greenbaum JA, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(Database issue):D405-D412.
Crossref - Sidney J, Assarsson E, Moore C, et al. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4(1):2.
Crossref - Dimitrov I, Flower DR, Doytchinova I. AllerTOP–a server for in silico prediction of allergens. BMC Bioinformatics. 2013;14(Suppl 6):S4.
Crossref - Gupta S, Kapoor P, Chaudhary K, et al. In Silico approach for predicting toxicity of peptides and proteins. PLOS ONE. 2013;8(9):e73957.
Crossref - Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531-552.
Crossref - Wang S, Peng J, Ma J, Xu J. Protein secondary structure prediction using deep convolutional neural fields. Sci Rep. 2016;18962.
Crossref - Schaarschmidt J, Monastyrskyy B, Kryshtafovych A, Bonvin A. Assessment of contact predictions in CASP12: co-evolution and deep learning coming of age. Proteins: Structure, Function, and Bioinformatics. 2017;86(Suppl 1):51-66.
Crossref - Yang Y, Gao J, Wang J, et al. Sixty-five years of the long march in protein secondary structure prediction: the final stretch? Brief Bioinform. 2018;19(3):482-494.
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725-738.
Crossref - Heo L, Shin W-H, Lee MS, Seok C. GalaxySite: ligand-binding-site prediction by using molecular docking. Nucleic Acids Research. 2014;42(W1):W210-W214.
Crossref - Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(suppl_2):W407-W10.
Crossref - Hebditch M, Carballo-Amador MA, Charonis S, Curtis R, Warwicker J. Protein-Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33(19):3098-100.
Crossref - Dombkowski AA, Sultana KZ, Craig DB. Protein disulfide engineering. FEBS Letters. 2014;588(2):206-2012.
Crossref - Kozakov D, Hall D, Xia B, et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12(2):255-278.
Crossref - Morla S, Makhija A, Kumar S. Synonymous codon usage pattern in glycoprotein gene of rabies virus. Gene. 2016;584(1):1-6.
Crossref - Forner M, Canas-Arranz R, Defaus S, et al. Peptide-Based Vaccines: Foot-and-Mouth Disease Virus, a Paradigm in Animal Health. Vaccines (Basel). 2021;9(5):477.
Crossref - Gopinath SM, Sumer S, Pranaya P. A strategy for the production of infective chimeric foot and mouth disease virus to develop vaccines for FMD. The Experiment. 2012;5(2).
- Segundo FD-S, Medina GN, Stenfeldt C, Arzt J, de Los Santos T. Foot-and-mouth disease vaccines. Vet Microbiol. 2017;206:102-112.
Crossref - de Villegas RMD, Seki C, Mattion NM, Konig GA. Functional and in silico Characterization of Neutralizing Interactions Between Antibodies and the Foot-and-Mouth Disease Virus Immunodominant Antigenic Site. Front Vet Sci. 2021;8:554383.
Crossref - Liao YC, Lin HH, Lin CH, Chung WB. Identification of cytotoxic t lymphocyte epitopes on swine viruses: multi-epitope design for universal T cell vaccine. PLoS ONE. 2013;8(12):e84443.
Crossref - Meza B, Ascencio F, Sierra-Beltran AP, Torres J, Angulo C. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: An in silico approach. Infection, Genetics and Evolution. 2017;49:309-317.
Crossref - Shey RA, Ghogomu SM, Esoh KK, Nebangwa ND, Shintouo CM, Nongley NF, et al. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci Rep. 2019;9(1): 4409.
- Hasan M, Ghosh PP, Azim KF, et al. Reverse vaccinology approach to design a novel multi-epitope subunit vaccine against avian influenza A (H7N9) virus. Microb Pathog. 2019;130:19-37.
Crossref - Ali M, Pandey R, Khatoon N, Narula A, Mishra A, Prajapati VK. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci Rep. 2017;7(1):9232.
Crossref - Pandey RK, Ojha R, Aathmanathan VS, Krishnan M, Prajapati VK. Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine. 2018;36(17):2262-2272.
Crossref - Scheiblhofer S, Laimer J, Machado Y, Weiss R, Thalhamer J. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev Vaccines. 2017;16(5):479-489.
Crossref - Berkmen M. Production of disulfide-bonded proteins in Escherichia coli. Protein Expr Purif. 2012;82(1):240-251.
Crossref - Kamel M, El-Sayed A, Castaneda Vazquez H. Foot-and-mouth disease vaccines: recent updates and future perspectives. Arch Virol. 2019;164(6):1501-1513.
Crossref - Gori A, Longhi R, Peri C, Colombo G. Peptides for immunological purposes: design, strategies and applications. Amino Acids. 2013;45(2):257-268.
Crossref - Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Bio technol Adv. 2012;30(5):1102-1107.
Crossref - Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.