ISSN: 0973-7510
E-ISSN: 2581-690X
Skin and soft tissue infections (SSTIs) are commonly occurring infections with mild to serious clinical manifestations. The incidence of wound sepsis in India ranges from 10-33%1,2. It is important to know the potential microbial pathogens causing wound infections for clinicians to start empirical treatment for patients, while laboratory culture reports are awaited. To identify the common microorganisms and their antimicrobial resistance pattern in pus samples. A total of 8656 pus samples were received in the Microbiology department from various OP and IP departments of Stanley Medical College Hospital, Chennai. The samples were processed in the laboratory for aerobic culture to isolate the pathogens and to perform antibiotic sensitivity testing as per standard protocol31. This prospective study was done for a period of twelve months (Jan 2018 to Dec.2018). Growth was observed in 5793 samples (66.92%), while growth was absent in 2863 samples (33.07%). Of the culture positive samples, 250 (4.31%) showed mixed infection, while 5543 samples (95.68%) yielded a single isolate. In this study, among the isolates (6043 in number), 5965 (98.70%) were bacterial and 78 (1.29%) were fungal. The most common bacterial isolate was Pseudomonas species(27.42%), followed by Staphylococcus aureus (15.60%), Klebsiella pneumoniae (11.95%), Escherichia coli (9.53%), Coagulase negative Staphylococci (9.22%) and Acinetobacter spp. (8.65%). Among the S.aureus isolates, 59% were Methicillin resistant and 41% were Methicillin sensitive. The fungal isolates were Candida spp. ( 80.76%) and Aspergillus spp. (19.24%). The common pathogens isolated in this study were Pseudomonas species (27.42%), Staphylococcus aureus (15.60%) and Klebsiella pneumoniae (11.95%). The increased incidence of antimicrobial-resistant microorganisms like Methicillin-resistant S. aureus, ESBL and MBL producers causes great global concern leading to more difficult to treat infections and death.
Antimicrobial Resistance, Skin and soft tissue infection, MRSA , ESBL, MBL
Skin and soft-tissue infections (SSTIs) encompass a variety of pathological conditions that involve the skin and underlying subcutaneous tissue, fascia, or muscle, ranging from simple superficial infections to severe necrotizing infections3. They are caused by microbial pathogens in wounds due to trauma, burns, and surgical procedures and result in the production of pus, a yellowish-white fluid formed as a part of an inflammatory response, composed of exudate containing dead WBCs, cellular debris, necrotic tissues and pathogenic bacteria.10. 26 Uncomplicated infections are often superficial and can be treated by incision and drainage alone or along with oral antibiotics The complicated SSTIs extend to subcutaneous tissue, fascia, or muscle and require a combination of antimicrobials with surgical intervention.29
According to the definitions of Centers for Disease control and Prevention, Criteria for Skin Infections include purulent drainage, pustules, vesicles or boils with tenderness, swelling or erythema. Criteria for Soft tissue infections include Purulent discharge from affected site, positive culture from tissue or drainage from affected site, abscess or gross evidence of infection. 30
Among hospitalized patients, the estimated prevalence of SSTIs is 7%–10%.29 In South India, incidence is about 2%2 and the mortality rate ranges from 4% (Singh et al, North India)8 to 14% (Abhilash et al, South India)2
Even with advances in infection control practices like improved operating room ventilation, sterilization methods, surgical technique and availability of antimicrobial prophylaxis, Surgical Site Infections (SSIs) remain a substantial cause of morbidity, prolonged hospitalization and mortality rate of 3% globally24. Surgical-site infection (SSI) is an infection of the skin or deep-space occurring at the incision or in the field of an invasive procedure within 30 days after operation (1 year for an implant) 24. SSIs are the most common healthcare-associated infection (HAI), accounting for 31% of all HAIs among hospitalized patients.29
Skin and soft tissue infections: open sores (ulcers, burns and bedsores) encourage bacterial colonization and may lead to systemic infection24. Pathogenic bacteria have greater virulence causing infections regardless of host status. Gram positive bacteria like Staphylococcus aureus (cutaneous bacteria that colonize the skin and nose of both hospital staff and patients) cause a wide variety of infections and are frequently resistant to antibiotics, especially MRSA (Methicillin-Resistant Staphylococcus aureus); beta-haemolytic Streptococci are also important as causes of skin and soft tissue infections.
Gram negative bacteria like Enterobacteriaceae (e.g. Escherichia coli, Klebsiella, Proteus, Enterobacter) may cause serious infections exhibiting resistance patterns like ESBL (Extended Spectrum Beta Lactamase) production and Carbapenem resistance (Carbapenem resistant Enterobacteriaceae, CRE). Other Gramnegative organisms implicated include Pseudomonas spp., known for survival in various disinfectants used in the hospital and carrying resistant genes23. More recently, multidrug-resistant Gramnegative bacteria–Acinetobacter baumannii, has become one of the commonest organisms causing SSTIs4,5,6, producing Metallobeta-lactamases (MBL) with resistance to Carbapenems, leaving a narrow choice of antibiotics for treatment.
Successful management of patients with severe SSTIs involves prompt recognition, appropriate antibiotic therapy, timely surgical debridement or drainage, and resuscitation when required3. Rapid emergence of antimicrobial resistant strains necessitates periodic evaluation of antimicrobial resistance patterns of potential pathogens to frame an antimicrobial policy for implementation in the health care setting.
The objective of this study is to identify the pyogenic bacteria from pus samples and to determine their antibiotic susceptibility to various antibiotics commonly used in therapy.
Study group
This is a cross-sectional study conducted at Stanley Medical College Hospital in Chennai, South India. In this study, patients of all age groups and both genders from out-patients and in-patients presenting with signs and symptoms of SSTI were included. Patients who were admitted in the hospital for more than 3 days, on prior antibiotic treatment, those with infected burns were excluded from this study. A total of 8656pus samples collected from out-patients and in-patients were studied during the period January 2018 to December 2018.
Specimen collection and processing
Number of pus samples collected from general surgery department were 3909, orthopaedics department 2555, medicine department 761, ENT department 543, other surgical specialities 487 and Obstetrics and gynaecology department 401.
Pus samples were collected using sterile cotton swabs placed in sterile tubes prepared and sterilized in-house by the lab and pus aspirates were collected using sterile disposable syringes (Paras syringes). Tissue specimens were obtained from wound margins, ulcers and deep-seated infections after surgical debridement of the wound and collected in Brain heart infusion broth.
Samples were immediately transported to the Microbiology laboratory and processed as per standard protocol.31Samples were inoculated on to Blood agar (BA) and MacConkey agar (MA) and the plates were incubated at 37°C for 24 to 48 hrs. Gram staining of the samples were done for microscopy. Tissues inoculated in BHI broth were subcultured after 24 hours. Bacterial culture isolates were identified by standard microbiological techniques like Gram staining and biochemical reactions such as catalase test, coagulase test, Urease test, Mannitol fermentation, Bile esculin hydrolysis and heat tolerance test for Gram positive cocci.
Gram staining, motility testing, Oxidase test, Indole test, Citrate utilization, Urease test, Triple sugar iron test, Mannitol motility medium test and phenyl pyruvic acid test were done for identification of gram negative bacilli.
Antibiotic Susceptibility testing
Antibiotic sensitivity testing of all isolates was done by Kirby Bauer’s disc diffusion method on Mueller Hinton agar and Zones of inhibition measured and results were interpreted after 18-24 hours as per Clinical Laboratory Standards Institute guidelines25. Mueller Hinton agar with 5% sheep blood was used for Antibiotic sensitivity testing of Streptococcal isolates.1st and 2nd line antibiotic discs were used for antibiotic sensitivity testing. If resistance was noted for these drugs, 3rd line of antibiotic discs were tested subsequently as per CLSI guidelines 25.1st and 2nd line of antibiotic discs used for Gram positive cocci were Ampicillin (10mcg), Penicillin(10 units), Erythromycin (15mcg), Clindamycin (2mcg), Cotrimoxazole (25mcg), Cefoxitin (30mcg), Linezolid(30mcg), Tetracycline (30mcg), Vancomycin (30mcg) and Cefotaxime (30mcg). 3rd line antibiotics used were Chloramphenicol (30mcg), Levofloxacin (5mcg), Ciprofloxacin (5mcg), Gentamicin (10mcg) and High level Gentamicin(120 mcg).
1st and 2nd line of antibiotic discs used for Gram negative bacilli were Ampicillin (10mcg), Amikacin (30mcg), Amoxy-clav (20/10mcg), Cefepime (30mcg), Ceftazidime (30mcg), Ciprofloxacin (5mcg), Levofloxacin(5mcg), Gentamicin (10mcg), Imipenem (10mcg), Meropenem(10mcg), Piperacillin/tazobactam (100/10mcg), Cotrimoxazole (25mcg), Aztreonam(30mcg). 3rd line antibiotics used were Choramphenicol (30mcg) and Tetracycline (30mcg).25
ESBL test
Extended spectrum Beta-lactamase production was determined by phenotypic double disc diffusion test. Lawn culture of the test organism was made on MHA. Antibiotic discs Ceftazidime (CAZ 30ìg) and Ceftazidime/ Clavulanic acid (CAZ/ CA 30ìg/ 10ìg ) were placed on the plate and incubated at 35°C overnight. An increase in zone diameter of 5mm or more for Ceftazidime/ Clavulanic acid compared to Ceftazidime alone confirmed an ESBL producing organism .25
MIC testing
To determine MIC values for Colistin & Tigecycline for Gram negative bacilli, Colistin 0.016-256 mcg/ml Ezy MIC strips (Himedia) and Tigecycline 0.016-256 mcg/ml Ezy MIC strips (Himedia) were used. MIC breakpoint for Colistin ≤ 2 is considered susceptible and >/=4 as resistant.25
The interpretative Tigecycline MIC susceptibility breakpoints of Enterobacteriaceae issued by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) is susceptible MIC, <1 µg/ml; resistant MIC, >2 µg/ml32
To determine MIC values for Vancomycin for Staphylococcus aureus, Vancomycin 0.016-256mcg/ml Ezy MIC strips (Himedia) were used.MIC breakpoint for Vancomycin is susceptible MIC, < 2 µg/ml and resistant MIC, >16 ìg/ml) 25
Quality Control
Quality control (QC)for antimicrobial susceptibility testing is done in the laboratory once a week and, in addition, every time a new batch of Mueller–Hinton agar or a new batch of discs is used. Performance testing of media used for culture and biochemical reactions are done once a week using control strains. The standard strains used for QC are Staphylococcus aureus (ATCC 25923), Enterococcus faecalis (ATCC 29212), Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853).27
Fungal culture
Fungal growth observed in Blood agar plates were sub-cultured onto Sabouraud Dextrose Agar and identified by colony characteristics, Germ tube test, Gram staining and Lactophenol Cotton blue mount.
Statistical Analysis
Data were collected and analyzed using SPSS software by descriptive statistical methods by computing means and proportion with 95% confident interval. A p < 0.05 was considered statistically significant.
A total of 8656 pus samples were collected and sent to the Microbiology laboratory for culture and antibiotic sensitivity testing. We received 7642 pus swabs (88.3 %), 961 aspirates (11.1%) and 53 tissue specimens (0.6%)
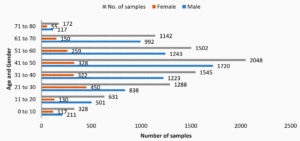
Males constituted the predominant population of the study group (6554, 75.7%), followed by females (1633, 18.9%) and paediatric age group (469, 5.41%). Many subjects were in the age group 41-50 years (23.66%), followed by 31-40 years (17.85%), 51-60 years (17.35%), 21-30 years (14.88%) and 61-70 years (13.19%) [Fig 1].
Maximum number of pus samples were sent from general surgery department (3909,45.15%), followed by orthopaedics department (2555, 29.51%), medicine department (761,8.79%), ENT department (543, 6.27%), other surgical specialities (487, 5.26%), Obstetrics and gynaecology department (401, 4.63%) [Fig.2].
Growth was observed in 5793 samples (66.92%)and absent in 2863 samples (33.07%). Of the growth positive samples, 250 (4.31%) were polymicrobial, whereas the remaining 5543 samples (95.68%) were monomicrobial.
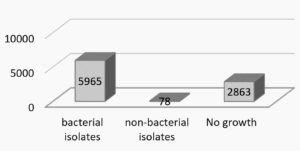
Among the 6043 isolates, 5965 (98.70%) were bacterial and 78 (1.29%) were fungal [Fig.3].
Among the 5965 bacterial isolates, the most common bacteria isolated was Pseudomonasspecies (1636 isolates, 27.42%), followed by Staphylococcus aureus (931, 15.60%), Klebsiella pneumoniae(713, 11.95%), Escherichia coli (569, 9.53%), Coagulase negative Staphylococci (550, 9.22%), Acinetobacter spp.(516, 8.65%), Proteus spp.(407, 6.82%), Klebsiella oxytoca (391, 6.55%), Enterococcus spp.(115, 1.92%), Streptococcus spp.(82, 1.37%) and Citrobacterspp.(55, 0.92%) [Fig 4].
Candida spp. (63, 80.76%) and Aspergillus spp. (15, 19.23%) were seen among 78 fungal isolates.Candida albicans was isolated in 13 patients (17%) ,Candida tropicalis in 27 patients (34%) ,other Non-albicans Candida species in 23 patients. (30%). Aspergillus flavus in 9 patients (11%) and Aspergillus fumigatus in 6 patients (7%).
Antimicrobial susceptibility test was carried out by Kirby Bauer Disc diffusion method as per Clinical Laboratory Standards Institute, M100 Performance standards for antimicrobial susceptibility testing.
Among Enterobacteriaceae, high sensitivity was noted to carbapenems (Imipenem and Meropenem) and Aztreonam. For the highly resistant isolates, E- MIC test was done with Colistin and Tigecycline Strips which showed 100% sensitivity [Table 1]. A proportion of 78% were detected to be ESBL producers and 20% as Carbapenem resistant enterobacteriaceae
Table (1):
Antimicrobial sensitivity of Gram negative bacilli.
Drugs |
Klebsiella pneumonia (713) |
Klebsiellaoxytoca (391) |
Escherichia coli (569) |
Proteus spp. (407) |
Citrobacter spp. (55) |
Acinetobacter spp. (516) |
Pseudomonas spp. (1636) |
|
|---|---|---|---|---|---|---|---|---|
Ampicillin |
IR |
IR |
4% |
– |
– |
– |
– |
|
Amoxycillin-Clavulanate |
16% |
16% |
15% |
– |
18% |
– |
– |
|
Cefotaxime |
20 % |
25% |
21% |
30% |
35% |
38% |
– |
|
Ceftazidime |
45% |
46% |
||||||
Gentamicin |
50% |
44% |
55% |
54% |
35% |
47% |
57% |
|
Amikacin |
62% |
60% |
77% |
72% |
65% |
67% |
79% |
|
Piperacillin-Tazobactam |
69% |
78% |
89% |
72% |
60% |
78% |
89% |
|
Cotrimoxazole |
46% |
45% |
49% |
40% |
25% |
47% |
IR |
|
Cefepime |
25% |
27% |
24% |
35% |
25% |
28% |
32% |
|
Imipenem |
79% |
86% |
90% |
86% |
80% |
85% |
92% |
|
Meropenem |
53% |
78% |
88% |
89% |
92% |
85% |
87% |
|
Ciprofloxacin |
37% |
25% |
40% |
48% |
28% |
46% |
50% |
|
Levofloxacin |
46% |
50% |
60% |
46% |
52% |
57% |
65% |
|
Tetracycline |
30% |
40% |
40% |
IR |
38% |
50% |
IR |
|
Aztreonam |
90% |
87% |
92% |
87% |
88% |
IR |
75% |
|
Tigecycline (E-MIC) |
100% |
100% |
100% |
IR |
100% |
100% |
IR |
|
Colistin (E-MIC) |
100% |
100% |
100% |
IR |
– |
100% |
100% |
- Not tested
Among non-fermenting Gram negative bacilli high sensitivity was noted for Carbapenems, Aztreonam as well as Amikacin. E-MIC test done for resistant isolates with Colistin and Tigecycline Strips showed 100% sensitivity[Table 1].
Gram Positive Cocci–92% of MSSA isolates were sensitive to Levofloxacin while only 78% of MRSA isolates were sensitive. Streptococci spp were sensitive to almost all antibiotics and 64 (78%) of the isolates were Streptococcus pyogenes. Linezolid was the consistently highly sensitive drug among all genera. Enterococci spp were highly sensitive to Vancomycin, Linezolid and Tigecycline. CoNS showed moderate sensitivity to many antibiotics. E-MIC test was done with Vancomycin strips for Staphylococcal isolates. [Table 2]
Table (2):
Antimicrobial sensitivity of Gram positive cocci.
Antibiotics |
MSSA (382) |
MRSA (549) |
CONS (550) |
Streptococci (82) |
Enterococci (115) |
|---|---|---|---|---|---|
Penicillin |
10% |
0% |
18% |
89% |
45% |
Erythromycin |
32% |
24% |
33% |
69% |
– |
Clindamycin |
44% |
39% |
42% |
94% |
– |
Gentamicin |
75% |
62% |
75% |
93% |
59% (HLG) |
Cotrimoxazole |
53% |
49% |
60% |
– |
– |
Levofloxacin |
92% |
78% |
85% |
100% |
– |
Ciprofloxacin |
– |
– |
– |
– |
58% |
Linezolid |
100% |
100% |
100% |
100% |
100% |
Cefepime |
– |
– |
– |
100% |
– |
Tetracycline |
– |
– |
– |
75% |
64% |
Vancomycin |
– |
100% (E-MIC) |
– |
100% |
100% |
Tigecycline |
– |
– |
– |
– |
100% |
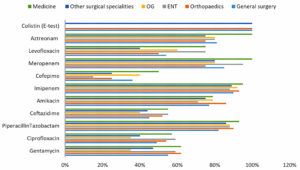
The isolates of Pseudomonas showed high sensitivity to Piperacillin-Tazobactam and Carbapenems in all wards.[Figure 5]
In this study,78% of Enterobacteriaceae were detected to be ESBL producers by double disc ESBL test. 59% of Staphylococcus aureus isolates were methicillin resistant .15% of isolates of Pseudomonas spp. and Acinetobacter spp. were MBL producers.
Skin and soft tissue infection is a common medical condition and the disease burden is high. In this study, a total of 8656 pus samples were collected from out-patients and in-patients with the aim of isolation of pathogenic micro-organisms and to study the antibiotic resistance pattern.
Table (3):
Resistance pattern among bacterial isolates.
Resistance |
No of isolates |
Percentage |
|---|---|---|
ESBL |
1665 |
78 % |
CRE |
427 |
20 % |
MBL |
323 |
15 % |
MRSA |
549 |
59 % |
Among the samples collected, 7642 were pus swabs (88.3 %) and 961 were aspirates (11.1%). This is similar to a study done by Upreti et al where 85.7% of the samples were pus swabs and 14.3% were aspirates.28
Pus samples were received maximally from general surgery department (3909, 45.15%), followed by orthopaedics department (2555, 29.51%). Higher number of samples from surgery department has been observed in many studies (Roopa et al)7. The most common age group affected by pyogenic infection in our study was 41-50 years and there was a predominance of males(75.7%) over female patients(18.9%). This was comparable to a study by Singh et al 8 and Roopa et al7.
Growth was observed in 5793 samples (66.92%). Isolation rate correlates with several studies done on pus cultures in developing countries like India and Nepal ranging between 60-75% (Esposito et al.14, Rai et al.9, Roopa et al.7, Trojan et al.10). Out of 5793 samples, 250 samples (4.3%) showed polymicrobial growth. Studies by Vijeta et al15 and Dhiraj Kumar et al16 show polymicrobial growth in 20% and 8.6% respectively. Open wounds get easily colonized and invaded by numerous bacteria as they provide a warm and moist environment for bacterial colonization and proliferation. This might be the reason for polymicrobial growth
About 6043 pathogens were isolated from various samples. Gram negative bacilli (GNB 71.8%) isolates were more compared to gram positive pathogens (28.13%). This is comparable to the study by Trojan et al10 where 77% of the isolates were GNB and that of Ioannou et al11 where 54.6% of the isolates were GNB.
Pseudomonas spp. (27.42%) was the commonest organism isolated followed by Staphylococcus aureus (15.60%), Klebsiella pneumoniae (11.95%) and E.coli (9.53%). Pseudomonas spp. was the commonest gram negative bacilli isolated by Esposito et al 14 and by Raytekar et al17 with isolation rate of 20.2%.
The number of fungal isolates were 78(1.29%) with Candida spp. accounting for 80.76% and Aspergillus spp. in 19.23%. Candida albicans was isolated in 17%, This was comparable to a study by Kaya D et al where 2% of the agents of surgical infections were yeasts: Candida albicans was isolated in 5 (55.6%)34
Antibiotic sensitivity testing of the isolated Gram negative bacilli showed that most of them were sensitive to Carbapenems, Piperacillin-tazobactam, Amikacin, Aztreonam and Levofloxacin but resistant to Ampicillin and Amoxycillin-Clavulanic acid. All the multidrug resistant strains were sensitive to Colistin and Tigecycline. These findings are correlating with several pus culture studies (Roopa et al.7).
Most of the isolates of Staphylococcus aureus and CoNS were highly sensitive to Linezolid, Levofloxacin and Gentamicin and moderately sensitive to Cotrimoxazole and Clindamycin. All the resistant strains were sensitive to Vancomycin. The Streptocooci isolates were highly sensitive to all the antibiotics. Enterococci spp were 100% sensitive to Linezolid and Vancomycin and moderately sensitive to Ciprofloxacin (58%) and High level Gentamicin (59%). In the study by Mudassar et al S.aureus isolates were highly sensitive to Gentamicin (86%)and Clindamycin (79%)33
Extended spectrum Beta-lactamase production as determined by double disk test was around 78%. This is comparable to a study done in India which gives ESBL rate of 68% by Agrawal et al18 and 84% by Perumal P.G et al19.
Carbapenem resistant Enterobacteriaceae (CRE) was 20% which is comparable with a nationwide review giving a rate of 10–20% (Hsu et al.12). MBL producing Non-fermenters (Pseudomonas spp., Acinetobacter spp.) was 15% which is comparable to the same study by Amandeep et al.13, which gives a prevalence of 21%.
Methicillin Resistant Staphylococcus aureus (MRSA) was isolated in 59% of Staphylococcus aureus strains which is in concordance with a study by Upreti et al (60%) and by Avinash Kumar et al21 (61%). The prevalence of MRSA in a study done by INSAR group, India20was 41% and 43% in the study performed by Haysom et al22.
In this study, Pseudomonas spp. was the most common pathogen isolated (27.42%)followed by Staphylococcus aureus(15.6%)%), Klebsiella pneumoniae (11.95%) and Escherichia coli (9.53%). The study indicates gram-negative bacteria as significant emerging causative agents of SSTI in our setting. Antibiotic susceptibility pattern of the isolated pathogens shows high sensitivity to higher antibiotics like Carbapenems, Piperacillin-Tazobactum and Linezolid. MBL production was observed in 15% of the Pseudomonas strains.78% of the Enterobacteriaceae isolates were ESBL producers and 59% of Staphylococcus aureus strains were methicillin resistant. This study shows the common pathogens causing Skin and soft tissue infections and their antibiotic sensitivity to plan empiric treatment for patients in our center.
Prolonged hospital stay, indiscriminate use of antibiotics and lack of awareness are the possible predisposing factors of emergence of ESBL and MRSA. This study highlights the importance of infection control practices in health care settings to prevent the spread of resistant strains and avoiding indiscriminate and irrational use of antibiotics.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Narula H, Chikara G, Gupta P. A prospective study on bacteriological profile and antibiogram of postoperative wound infections in a tertiary care hospital in Western Rajasthan. J Family Med Prim Care. 2020;9:1927-34.
Crossref - Abhilash KP, Varghese S. Profile and outcome of patients presenting with skin and soft-tissue infections to the emergency department. Curr Med Issues. 2019:17-30-3.
Crossref - Sartelli M, Guirao X, Hardcastle TC, et al. 2018 WSES/SIS-E consensus conference: Recommendations for the management of skin and soft-tissue infections. World J Emerg Surg. 2018; 13:58.
Crossref - Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939-51. PMID: 18007677.
Crossref - Leung WS, Chu CM, Tsang KY, Lo FH, Lo KF, Ho PL. Fulminant community-acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest. 2006;129(1):102-9. PMID: 16424419.
Crossref - Sunenshine RH, Wright MO, Maragakis LL, et al. Multidrug-resistant Acinetobacter infection mortality rate and length of hospitalization. Emerg Infect Dis. 2007;13(1):97-103.
Crossref - Roopa C, Deepali V, PUS culture isolates and their antibiotic sensitivity at a Tertiary Care Hospital in Hyderabad Karnataka Region. IP Int J Med Microbiol Trop Dis. 2017;3(4):140-145
- Singh B, Singh S, Khichy S, Ghatge A. Clinical Presentation of Soft-tissue Infections and its Management: A Study of 100 Cases. Niger J Surg. 2017;23(2):86-91.
Crossref - Rai S, Yadav UN, Pant ND, et al. Bacteriological Profile and Antimicrobial Susceptibility Patterns of Bacteria Isolated from Pus/Wound Swab Samples from Children Attending a Tertiary Care Hospital in Kathmandu, Nepal. International Journal of Microbiology. 2017;2017:2529085.
Crossref - Trojan R, Razdan L, Singh N. Antibiotic Susceptibility Patterns of Bacterial Isolates from Pus Samples in a Tertiary Care Hospital of Punjab, India. International Journal of Microbiology. 2016;2016:9302692.
Crossref - Ioannou P, Tsagkaraki E, Athanasaki A, Tsioutis C, Gikas A. Gram-negative bacteria as emerging pathogens affecting mortality in skin and soft tissue infections. Hippokratia. 2018; 22(1):23-28.
- Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, Tambyah PA. Carbapenem-Resistant Acinetobacterbaumannii and Enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev. 2016;30(1):1-22.
Crossref - Kaur A, Singh S, Prevalence of Extended Spectrum Betalactamase (ESBL) and Metallobetalactamase (MBL) Producing Pseudomonas aeruginosa and Acinetobacterbaumannii Isolated from Various Clinical Samples, Journal of Pathogens.2018;2018:6845985.
Crossref - E Silvano, DS Giuseppe, P Angelo, et al. Epidemiology and microbiology of skin and soft tissue infections: preliminary results of a National registry, Journal of Chemotherapy. 2019;31(1):9-14.
Crossref - Sharma V, Parihar G, Sharma V, Sharma H.A Study of Various Isolates from Pus Sample with Their Antibiogram from Jln Hospital, Ajmer. IOSR Journal of Dental and Medical Sciences (IOSR-JDMS). 2015;14(10):64-68.
- P Pokhrel, A Shrestha, P Panthi, S Manadhar, DK Chaudhary. Bacteriological Profile and Antibiotic Susceptibility Pattern of Wound Infection in Children. EC Microbiology. 2017;5(3):93-100.
- Raytekar NA, Choudhari MR, Das S. Antibiotic profiling of Pseudomonas aeruginosa isolates from pus sample of rural tertiary care hospital of Western. Maharashtra, Loni, India. Int J Res Med Sci. 2017;5:3076-81.
Crossref - Agrawal SK, Panigrahy A, Perumalla S, Kapil A, Dhawan B. Microbiological profile and antibiotic resistance pattern of skin and soft-tissue infections: A study from Northern India. J Lab Physicians. 2018;10(4):471-472.
Crossref - Perumal PG, Jnaneshwara KB, Patil AB, Akshay R. Incidence of infections with extended spectrum beta Lactamase (ESBL)-producing gram-negative bacteria among patients admitted in medical intensive care unit of tertiary care hospital. Trop J Path Micro. 2017;3(2):168-173.
Crossref - Indian Network for Surveillance of Antimicrobial Resistance (INSAR) group, India. Methicillin resistant Staphylococcus aureus (MRSA) in India: prevalence & susceptibility pattern. Indian J Med Res. 2013;137(2):363-369.
- Kumar A, Kumar A. Prevalence of Methicillin Resistant Staphylococcus Aureus (MRSA) In A Secondary Care Hospital In North Eastern Part of India. Archives of Infect Diseases & Therapy. 2018;2(1).
Crossref - Haysom L, Cross M, Anastasas R, Hampton S, Harris M, Sneddon K. Methicillin-resistant Staphylococcus aureus skin and soft tissue infections in young people in custody in New South Wales. J Paediatr Child Health. 2019;55(2):224-228. PMID: 30161281.
Crossref - Prevention of Hospital acquired infection, A practical guide, 2nd edition; WHO manual; 2012.
- NHSN Patient Safety Component manual; CDC; 2017.
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
- Ki V, Rotstein C. Bacterial skin and soft tissue infections in adults: A review of their epidemiology, pathogenesis, diagnosis, treatment and site of care. Can J Infect Dis Med Microbiol. 2008;19(2):173-184.
Crossref - Jozef V, Kraesten E, Rohner P, et al. Basic laboratory procedures in clinical bacteriology / J. Vandepitte … [et al.], 2nd ed. World Health Organization. 2003. https://apps.who.int/iris/handle/10665/42696
- Upreti N, Rayamajhee B, Sherchan S, et al. Prevalence of methicillin resistant Staphylococcus aureus, multidrug resistant and extended spectrum â-lactamase producing gram negative bacilli causing wound infections at a tertiary care hospital of Nepal. Antimicrob Resist Infect Control. 2018; 7:121.
Crossref - Cardona AF, Wilson SE. Skin and Soft-Tissue Infections: A Critical Review and the Role of Telavancin in Their Treatment. Clinical Infectious Diseases. 2015;61(suppl_2): S69-S78.
Crossref - Centers for Disease Control and Prevention CDC/NHSN surveillance definitions for specific types of infections 2021. http://www.cdc.gov/nhsn/PDFs/pscManual/17pscNosInfDef_current.pdf
- Collee JG, Mackie T, Elvins J. Mackie and McCartney Practical Medical Microbiology McCartney 14th Edition 1996.
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. January 2017.
- Mudassar S, Khan SW, Ali M, Mahmood F. Aerobic Bacteriological Profile and Antimicrobial Susceptibility Pattern of Pus isolates in a Teaching Hospital, Lahore, Pakistan. International Journal of Contemporary Medical Research. 2018; 5(4).
Crossref - Kaya D, AldirmazAgartan C, Yucel M. Fungal Agents as a Cause of Surgical Wound Infections: An Overview of Host Factors. Wounds. 2007;19(8):218-22. PMID: 26110365.
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.