ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.29 | © The Author(s). 2019
Biogenic amines (BAs) have attracted increasing attention due to their significance in food safety and quality. The present study aimed at isolation of BAs producing microorganisms from different fermented products, namely kanji, sauerkraut, fish pickle and meat pickle and isolation of BAs degrading bacteria from canine saliva. A total of 63 BAs producing organisms identified among 356 organisms. Initial screening of BAs producing strains was carried out using Moller’s decarboxylase media and 2 BAs degrading organisms identified among 28 organisms. BAs presence is qualitative and quantitative confirmation by TLC and HPLC, respectively, using dansyl derivatized. They were characterized biochemically and identifying using 16S rDNA gene sequencing as Enterobacter hormaechei (18), Enterobacter cloacae (12), Bacillus licheniformis (14), Morganella morganii (11), Lactobacillus sakei (08) and Lactobacillus plantarum (2). Among all isolates, member of the Enterobacteriaceae family yielded the highest amount of putrescine (61.0-244 mg/100 gm), cadaverine (15.8-154 mg/100 gm), histamine (50-210 mg/100 gm) and least amount of tyramine (3-17.2) followed by Bacillus licheniformis which produced putrescine (58.3 mg/100 gm), cadaverine (83.3 mg/100 gm) and histamine (31.7 mg/100 gm). Lactobacillus sakei produced the highest amount of tyramine (21.2 mg/100 gm) and Lactobacillus plantarum from canine saliva degrade all important BAs histamine (100%), tyramine (100%), cadaverine (28%) and putrescine (67.8%). The result of the present study suggests that the BAs producing organisms are frequently present in fermented products, therefore, may pose a potential risk to human health and Lactobacillus plantarum from canine saliva applied as a potential BAs degradation bacterium in foods.
Biogenic amines, HPLC, Screening, Fermented foods, biogenic amines degradation, canine saliva, Lactic acid bacteria, Enterobacteriaceae.
Fermented foods and beverages are the vital components of the dietary culture widespread in nature around the world. When the fermentation process takes place, microbial growth, proteolysis and acidification provide favorable conditions for BAs production (Hugas et al. 1993). BAs are low molecular weight organic bases with aromatic (tyramine and phenylethy-lamine), heterocyclic (histamine and tryptamine) and aliphatic structure (putrescine, spermine, spermidine and cadaverine), synthesized by the bacterial decarboxylation of precursor amino acids in foods and beverages (Moracanina et al. 2015).
BAs are commonly present in the normal metabolism of humans, animals, plants and microorganisms at notable degree in protein rich foods like fish, fish products, meat products, fermented sausages, vegetables, soy products, dairy products like cheese, cider, fruits, nuts, chocolates and alcoholic beverages such as wine and beer as a consequence of microbial activity (ײnal. 2007). Amount of BAs rapidly increases with the increase in microbial growth and decarboxylase activity in the products. Microbial species present in these foodstuffs mainly lead to the accumulation of histamine, tyramine, cadaverine, tryptamine, putrescine and 2-phenylethylamine (Gardini et al. 2016).
The commonly observed food borne intoxication among BAs is caused by histamine. Consumption of the increased amount of BAs is found to be involved in food poisoning. A hazard level of BAs for human health has been propounded as 500 mg/kg of histamine, 100-800 mg/kg of tyramine and 30 mg/kg of phenylethy-lamine, putrescine and cadaverine do not show direct toxic effects however, they can enhance the toxic effects of other amines (Mah et al. 2003).
BAs also lead to several toxicological effects such as respiratory distress, headaches, hyper or hypotension, heart palpitation and several allergic disorders and they are potential precursors of carcinogenic nitrosamines, owing to the existence of a nitrosable factor in many foods. Besides this, BAs cause death in human when consumed at higher doses (ten Brink et al. 1990). Because of all these the effects degradation is a foremost aspect among this study. To limit the microbial growth, numbers of planned technologies have been designed like hydrostatic pressure, freezing, irradiation, controlled atmosphere packaging and use of food additives, but this method of application cannot eliminate already abide BAs since it is heat stable in nature (Luten et al. 1992). As a consequence of this reason, using microorganisms to degrade BAs has become an emerging method (Naila et al. 2010). Apart from these toxicological conditions, BAs are also related in food hygiene and their presence in high levels is indicative of a spoiling process (Vidal-Carou et al. 1990). Even the regulatory aspects concerning their qualitative and quantitative presence in various foods have been clearly worked out.
Despite, these BAs producing organisms, established to be a health damaging and spoilage causing microorganisms. The variability of microorganisms to decarboxylate amino acids and it is degradation is often being strain specific so their characterization demands investigation of different strain of the BAs producing and degrading species. The present study aims to determine the ability of different microorganisms in the production of BAs such as histamine, tyramine, cadaverine and putrescine isolated from different natural fermented products and degradation of BAs by microorganisms isolated from canine saliva, Among all pet animals canine are humans inimitable friend therefore canine isolate selected over other pet animal for the research. Even it is having a number of beneficial microorganisms to human (Ranjekar et al. 2015), they are characterized by biochemical and molecular method. To best of our knowledge there is no literature data on BAs degrading bacteria from canine saliva and this is first report on this study. BAs producers and degrading isolates were screened and characterized by 16s rDNA sequencing. Qualitative and quantitative confirmation of BAs production was done by Thin Layer Chromatography (TLC) and High Pressure Liquid Chromatography (HPLC) methods, respectively (Moracanina et al. 2015).
Chemicals
All microbiological media and chemicals were procured from Himedia, Mumbai (India). Molecular biology reagents and BAs standards histamine dihydrochloride, tyramine hydrochloride, cadaverine dihydrochloride, putrescine dihydrochloride, amino acid / precursor L-histidine, L-lysine, L-ornithine and L-tyrosine and dansyl chloride were obtained from Sigma-Aldrich (India). Acetonitrile and water (HPLC grade) were purchased from SRL (India).
Bacterial strains and Growth Conditions
The cultures isolated from different fermented foods and canine saliva isolates collected from MB division DFRL, the LAB strains were grown in the de Man Rogosa Sharpe (MRS) broth and the non-LAB strains were grown in Brain-heart infusion medium (BHI) broth respectively at 37°C for 24 h and store at 4°C.
Screening of BAs Producing Bacteria
A total of 356 isolates screened from different fermented food products (Table 1) and were grown in MRS and BHI broth at 37°C for 24-48 h. Production of BAs from each isolate was determined using the Moller’s decarboxylase agar (MDA) and Moller’s decarboxylase base broth (MDB) (pH 5.3) containing 0.05% precursor such as L-histidine, L-tyrosine, L-lysine and L-ornithine (Arribas et al. 2003). The screened isolates were inoculated to the MDA and MDB with and without precursor (as control) and were incubated at 37°C for 4 days and 7 days respectively. The color of Moller’s decarboxylase media changed from yellow to purple after incubation which indicates positive for BAs production, whereas the media remained yellow indicates negative for BAs production.
 Degradation Experiment
Degradation Experiment
A total of 28 isolates screened from canine saliva were grown in MRS and BHI broth at 37 °C for 24-48 h, then centrifuged at 12000 rpm for 10min at 4°C to separate the cells from medium, collect the pellet and discard the supernatant and cells were washed using sterile phosphate buffer 0.03M (NaH2PO4 13.8 gm/L, Na2HPO4 26.8 gm/L; pH 7.0) and resuspended in the same with 0.25 mM of BAs (histamine, tyramine, cadaverine and putrescine), as separately or as a mixture in the ratio 1:10 to obtain an optical density of 5.0 at 600 nm. Twenty ml of this cell suspension was transferred into sterile 100 ml Erlenmeyer flask and incubated with shaking at 37°C for 24-48 h (Bהumlisberger et al. 2015).
Biochemical Characterization
The BAs producing and degrading isolates were characterized according to FDA, 1992. Gram stain, motility, catalase test, oxidase test, indole test, methyl red test, Voges-Proskauer test, citrate test, urease test, H2S production test, ortho-Nitrophenyl-ג-galactoside (ONPG) test, nitrate reductase test, amino acid decarboxy-lation test like histidine decarboxylase (HDC), tyrosine decarboxylase (TDC), ornithine decarboxylase (ODC), lysine decarboxylase test (LDC), gelatin hydrolysis test, Deoxyribonuclease (DNase) test, esculinase test, fermentation of carbohydrates like D-glucose, lactose, maltose, L-rhamnose, sucrose, L-arabinose, xylose, melibiose, cellobiose, D-mannitol, glycerol, raffinose, D-sorbitol, gas production from glucose using durham’s tubes.
Qualitative analysis of BAs producing and degrading bacteria by TLC
Preparation of Standard Amines
A stock standard solution histamine, tyramine, cadaverine and putrescine were prepared by dissolving accurately 0.20–0.25 gm of each amine in 5% (w/v) trichloroacetic acid (TCA) solution (10ml). A 10-fold dilution of prepared solution formed the working standard.
Derivatization of the Amines
The amines were derivatized following the method of Shakila et al. (2001). Working standard solution (1.0 ml) was taken in an amber screw cap test tube, to which, 1.0 ml of phosphate buffer (pH 9.0), a drop of 4N sodium hydroxide solution and 2.0 ml of dansyl reagent (50 mg of dansyl chloride in 10 ml of acetone) was added. After thorough mixing the tubes were incubated at 55 °C for 1 h for dansylation. Similarly, the amines produced by the positive isolates were derivatized using the same protocol. Standards and samples were stored at 4 ± 2°C until further use.
TLC Method
Dansylated standard amines and samples were analyzed qualitatively using TLC (Shakila et al. 2001). Amines were fractioned (10µl) on pre-coated silica gel GF254 TLC plate (0.25 mm thickness, 5׳20 and 20׳20, E.Merck, India), and developed using chloroform: triethylamine (100: 25) mixture. The plate was sprayed with isopropanol: triethanolamine (8: 2) to enhance the fluorescence. Fractioned fluorescent amine spots were detected under the UV light at 365 nm (Scientific sales syndicates, Bombay, India).
Quantitative Analysis of BAs Production and Degradation by Bacteria using HPLC
Preparation of Standard Amines
Standard amines were prepared according to method of Chen et al. (2010). The concentration of each amine was 1 mg/ml, which was prepared by dissolving histamine dihydrochloride (82.8 mg), tyramine hydro-chloride (63.4 mg), cadaverine dihydrochloride (85.7 mg) and putrescine dihydrochloride (91.5 mg) in 50 ml of a 0.1 M HCl.
Derivatization of the Amines
A known concentration of (1-5mg) of each amine (1ml each) was taken in amber screw cap tubes. To this 0.2 ml of 2 M sodium hydroxide, 0.3 ml of saturated sodium bicarbonate and 2 ml of 1% dansyl chloride reagent (1% of dansyl chloride in 10 ml acetone) was added after thorough mixing the tubes and were incubated at 40 °C for 45 min. After the reaction, 100µl of ammonia was added and allowed to stand for 30 min at room temperature. Acetonitrile was added to make up volume to 5 ml and the solution was centrifuged at 10,000 rpm, at 4°C for 10 min. The supernatant was filtered (millipore) through 0.45 µm filter, and then used for HPLC for calibration of standard amines.
Sample Preparation and BAs Analysis
The amines producing the isolates and BAs degrading isolates were derivatized by taking the culture supernatant (centrifuged at 1000 rpm for 10 min). An equal volume of 5% TCA was added and filtered through Whatman No.2 filter paper. Filtrates were derivatized with dansyl chloride using the same procedure as per the dansylation of the standard amines.
The BAs produced by the bacteria chromatographicly analyzed using an Agilent technologies model 1260 USA, equipped with an auto injector, Photo Diode Array (PDA) detector and Chem Station software and chromatographic analysis was carried out for BAs degradation by bacteria using JASCO, HPLC system equipped with quaternary pump, Rheodyne injector, MD-2010 plus multiwave length detector with JASCO LC-Net I/ADC software. Separation of BAs was achieved using RP-C18 column (250 mm ׳ 4.6 mm i.d and 5.0 µm particle size column) and the mobile phase composing of acetonitrile (A) and water (B). Mobile phase was degassed and filtered through 0.22mm filters under vacuum before delivering into the system, The linear gradient programme was optimized by the percentage change of mobile phases and set as follows: Ttime/mobile phase-A: B (%): T0-50: 50; T19-90: 10; T20-90: 10; T30-50: 50; T35-50: 50 with constant flow rate of 1.0 ml/min. Injection volume used was 20 µl and kept the column temperature and sample cooler temperature at 25 ±2°C. The analytes were detected in PDA at 254 nm and amine separation time of 35 min was required for each sample with equilibrated column (Chen et al. 2010).
Genotypic Characterization of BAs Producing and Degrading Bacteria
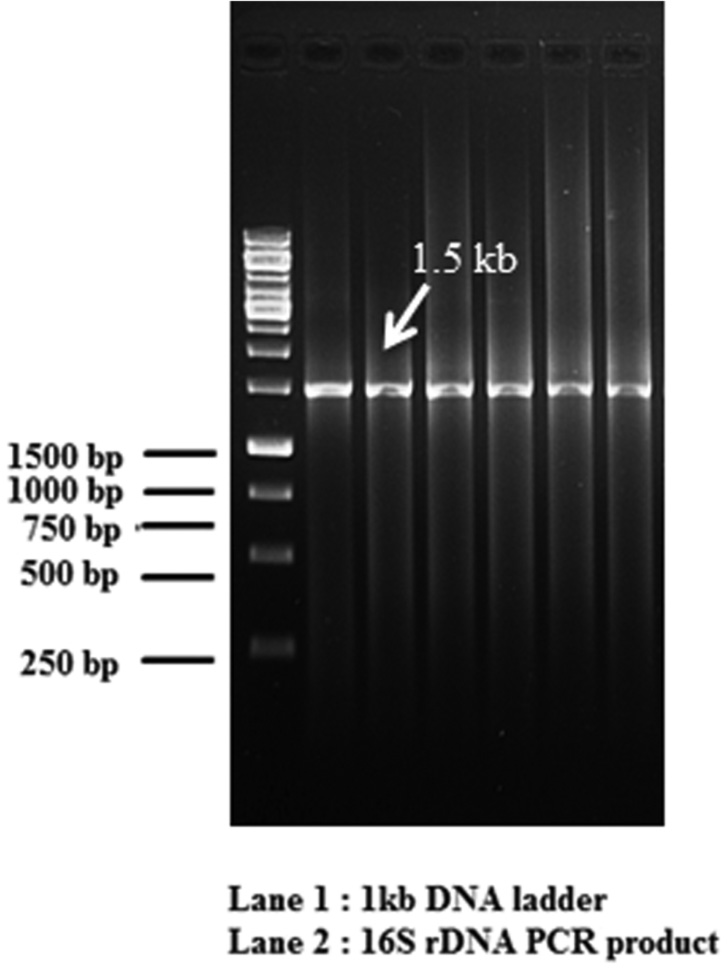
Total genomic DNA (gDNA) from bacterial cultures was isolated according to a protocol described by Sambrook et al. (1989). DNA precipitate was pelleted at 10,000 rpm and washed with 70% ethanol, air-dried and resuspended in 50 µl of 10 mM Tris-HCl. DNA sample prepared was quantified using Nanodrop 2000 spectrophotometer (Thermo Scientific Nanodrop, USA), and stored at -20°C until use. 16S rDNA of bacterial cultures was amplified using conventionally purified gDNA using the PCR primers P3 (forward primer) 5’-AGAGTTTGA-TCATGGCTCAG-3’ and P13 (reverse primer) 5’-GGTTACCTTGTTACGACTT- 3’ Weisburg et al. (1991). PCR reaction was performed in a micro centrifuge tubes using template DNA, PCR Buffer (1x), MgCl2 (2.0 mM), dNTPs (deoxynucleotide triphosphate) (50 µM), P3 primer (0.6 pmol), P13 primer (0.6 pmol), Taq polymerase enzyme (1.25 units). The reaction was performed in a thermal cycler (Veriti, Applied Biosystems, USA) using the following cycling parameters: Initial denaturation 4 min at 94°C, followed by 30 cycles of 1 min at denaturation 30 sec at 94°C, annealing 30 sec at 58°C, extension 45 sec at 72°C final extension 8 min at 72°C. Amplified products were resolved on a 1% agarose gel. The amplifications products were purified on QIAquick spin Columns (Qiagen, Germany) for direct sequencing. DNA sequencing was carried out using an ABI Genetic analyser 3500 DNA sequencer (Applied Biosystems, USA). The sequences were analysed with the BLAST (Basic Local Alignment Search Tool) the National Center of Biotechnology Information (NCBI) for identification of BAs forming and degrading bacteria and the best match of resemblance was selected to identify the bacterium.
BAs Produced Isolates and its Molecular Identification
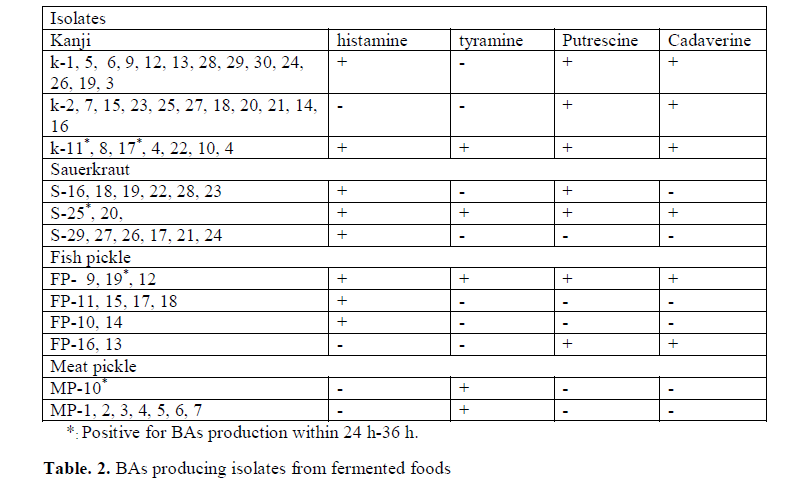
In the study 356 strains were randomly selected from 12 different fermented products mentioned in (Table 1), investigated for their potential to produce BAs (histamine, tyramine, cadaverine and putrescine). Out of 356 strains, 63 strains were positive for BAs and the number of putative strains for different BAs production is shown in (Table 2). The strains K (1-30), S (16-29), FP (9-19) and MP (1-7 and 10) obtained from kanji (A fermented beetroot beverage), sauerkraut, fish pickle and meat pickle, respectively displayed development of purple color in MDM, indicative of BAs production. The BAs producing organisms normally have been reported from fish, meat and sauerkraut (Taylor and Prester 2011). In the present investigation, BAs producing micro-organisms from kanji is reported for the first time. The strains S20, S25 recovered from sauerkraut, though produced histamine, cadaverine and putrescine but were negative for tyramine. At the same time, when tyrosine is used as precursor, it did produced purple color in the media, which otherwise provided false result, confirmed by TLC and HPLC. The change in color could be due to the alkaline products of bacterial origin, which turns media to purple color (Marcobal et al. 2006).
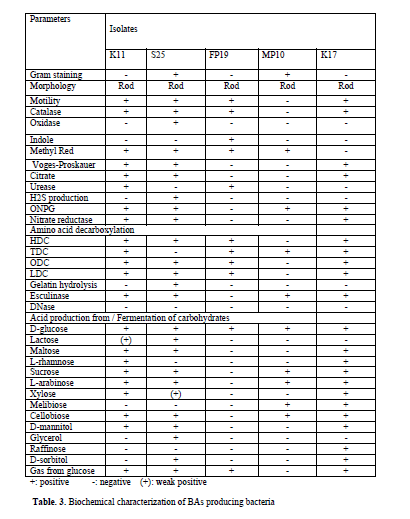
Though it is established that the fermented foods like sauerkraut, fish pickle and meat pickle can yield BAs producing strains belonging to different species and genera, quantity and types of BAs also stay varied. In our investigation as well, this variability was observed amongst different strains, recovered from the fermented food products studied. The strains (K11, K17, S25, FP19 and MP10) exhibiting fast development of color ( 24-36 h) and with high intensity of color (Fig. 1) were assumed to be having reasonably high level of BAs production, and they were characterized biochemically (Table 3) and were further processed for molecular identification (Fig. 3) and characterization.
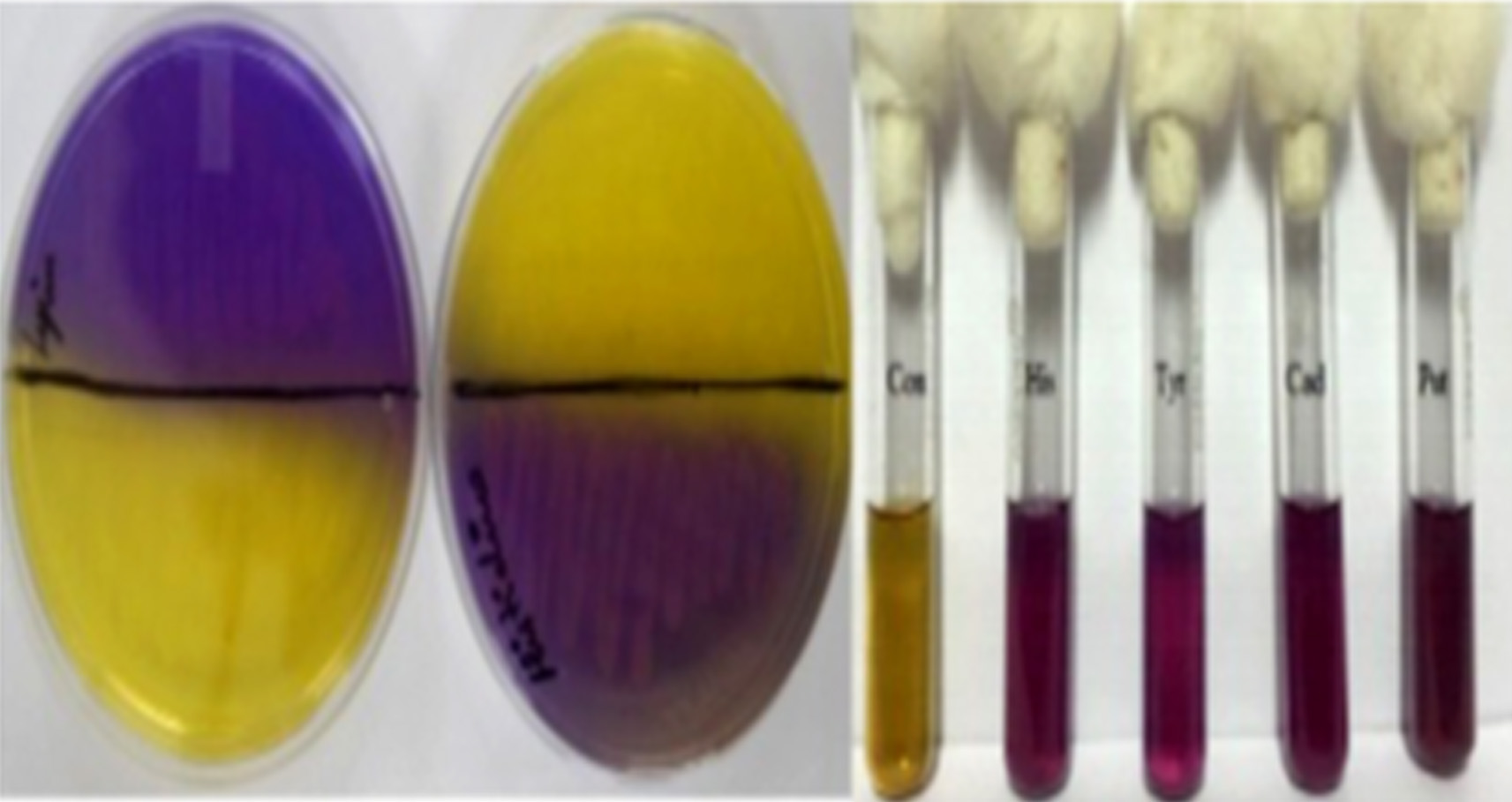
Fig. 1.Isolates showing positive for BAs production in MDA and MDB Con: Control. His: Histamine; Tyr: Tyramine; Cad: Cadaverine; Put: Putrescine
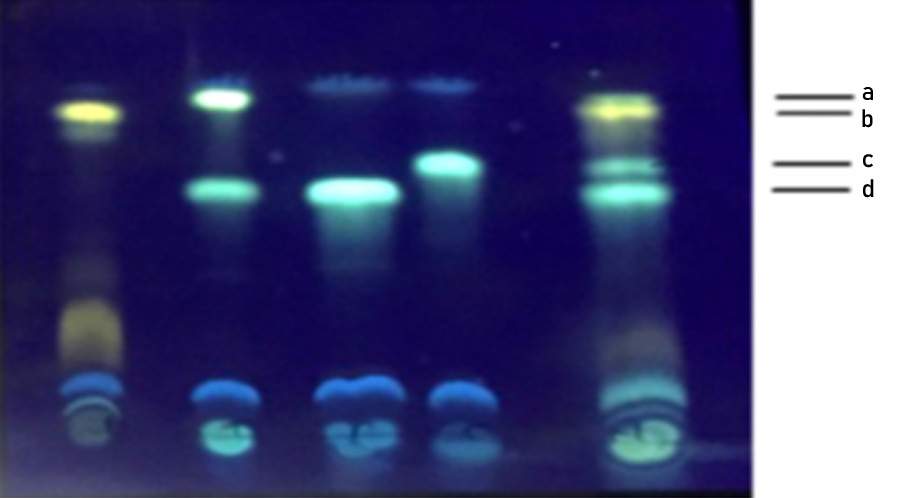
Fig. 2. TLC separation of dansylated standard amines and the amines produced by E. hormaechei on a precoated silica gel plate. Lane 1: Histamine, Lane 2: Tyramine and Putrescine, Lane 3: Putrescine, Lane 4: Cadaverine and Lane 5: Standards a: Tyramine, b: Histamine, c: Cadaverine, d: Putrescine
BLAST analysis of 16S rDNA gene sequence of the selected strains showed alignment of the tested strain with the reported 16S rDNA sequence of established organisms as evidenced from the available from the gene bank data. From the result obtained, the strains K11, K17, S25, FP19 and MP10 were identified as Enterobacter hormaechei, Enterobacter cloacae, Bacillus licheniformis, Morganella morganii and Lactobacillus sakei respectively.
BAs Degraded Isolates and its Molecular Identification
In degradation studies 28 isolates which were collected from MB division DFRL, the LAB strains were grown in the de Man Rogosa Sharpe (MRS) broth and the non-LAB strains were grown in Brain-heart infusion medium (BHI) broth respectively along with standard histamine, tyramine, cadaverine and putrescine. After 24h of incubation, sample was derivatized with dansyl chloride and subjected to further qualitative and quantitative analysis by TLC and HPLC. Initially presence and absence of BAs were detected by TLC where band formation from isolates was compared with standards BAs. BAs degradation amount qulitatively detected by HPLC method, where two isolates D18 and D21 showed ability to degrade 100% of histamine and tyramine, 28% of putrescine and 67.8% of cadaverine as shown in (Fig 9) Hence these two D18 and D21 strains were selected for further degradation studies because of high degradation capability based on HPLC results. The selected BAs degrading strains were analyzed by BLAST analysis of 16S rDNA gene sequence with the reported 16S rDNA sequence of established organisms as evidenced from the available data from the gene bank. From the result obtained, the D18 and D21 were identified as L. plantarum but this is first time reported that canine saliva isolate canine saliva proved to be significant BAs degrader as compared to other reported strain of L. plantarum in previous studied from different sources (Capozzi et al. 2012).
Qualitative Analysis of BAs Produced and Degraded Bacteria by TLC
TLC is used routinely as a rapid, qualitative method for the analysis of BAs. All the BAs producing bacteria namely E. hormaechei, E. cloacae B. licheniformis, M. morganii, L. sakei and BAs degrading bacteria L.planatrum showed constant Rf values of histamine (Rf 0.87), tyramine (Rf 0.911), cadaverine (Rf 0.724) and putrescine (Rf 0.617) chromatographic separation in TLC (Fig 2) shows the TLC separation of different BAs correspond to standard BAs. The precoated TLC plate was found to offer a neat and a good reproducible resolution of different amines based on increasing molecular weight. BAs bands detected under the UV light at 365 nm showed fluorescent spots of histamine appeared as yellowish, tyramine as green, putrescine and cadaverine as greenish blue spots (Shakila et al. 2001).
In degradation study the histamine and tyramine band in TLC were absent, indicating complete degradation of that amine and faint bands of cadaverine and putrescine is partially degraded in comparison to a standard BAs.
Since amines are highly polar substance that tends to streak during chromatography development and limiting the resolution, dansyl chlorides are used for derivatization of amines (Mo Dugo et al. 2006), which results in easy detection at a very low concentration owing to their fluorescent characteristics. Dansyl chlorides react with both primary and secondary amino groups, thereby producing highly stable fluorescent derivatives (Loukou and Zotou 2003). During the process of derivitization using dansylchloride, amino acids and ammonia present in the prepartion to get derivitized but due to their lower mobility remain near the origin and do not interfere with the separation of amines. Earlier literature suggests, TCA yields almost good recovery of amines and this recovery is also rapid in and around 2 min when used TCA at 5% concentration (Shakila et al. 2001). Based upon these references we could also observe rapid recovery of amines employing TCA.
Quantitative Analysis of BAs Produced and Degraded by Bacteria using HPLC
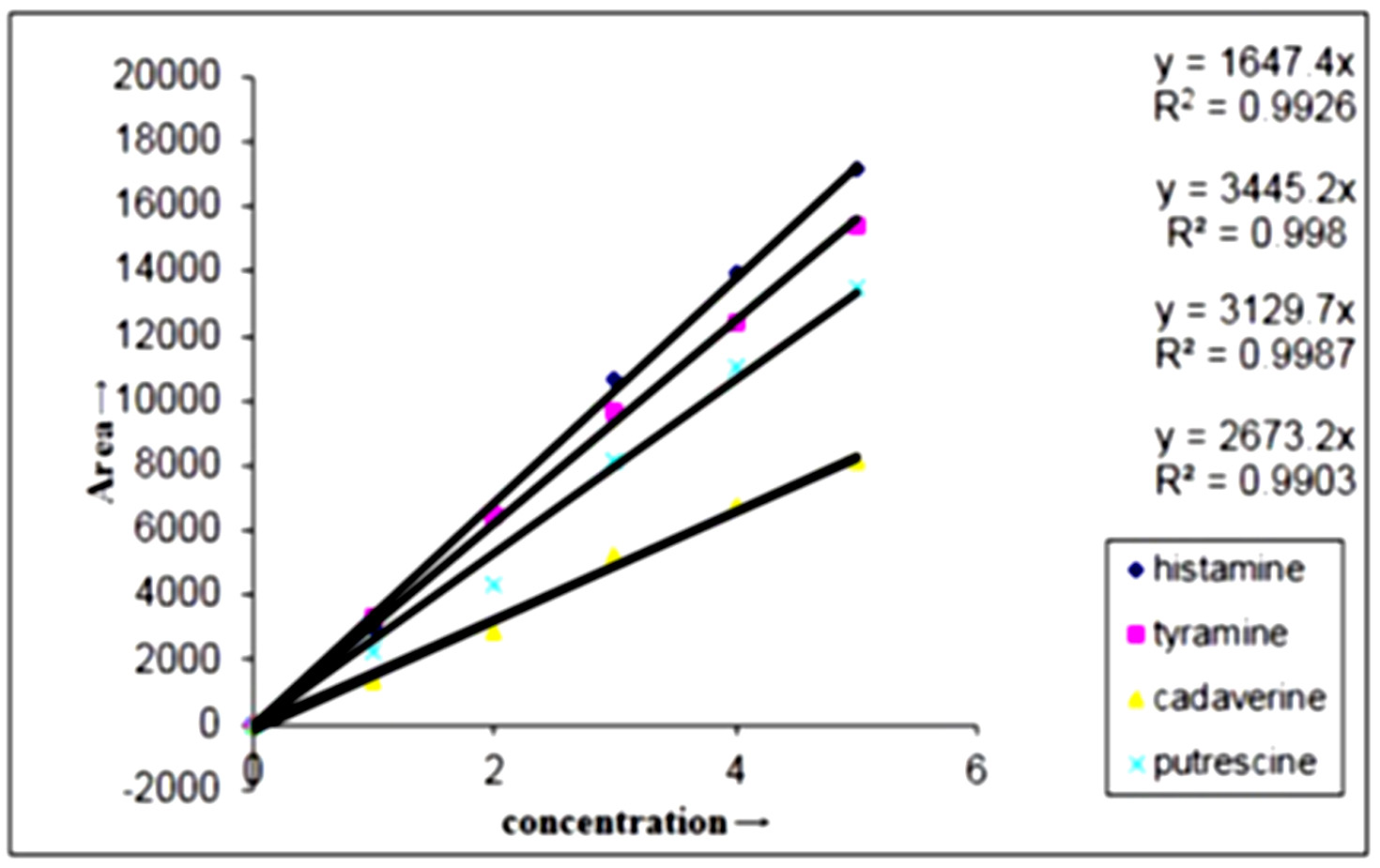
The BAs produced by E. hormaechei, E. cloacae, B. licheniformis, M. morganii and L. sakei determined quantitatively by HPLC is depicted in (Fig. 6) (Chromatograph of E. cloacae) and BAs degradation by L. plantarum determined quantitatively by HPLC is depicted in (Fig. 8) and all four amines that were derivatized with dansyl chloride got separated in 30 min with good resolution, sharpness and symmetry (Fig 5 and Fig. 7). Standard curves of four amines prepared separately in the range of 0-20 µg/ml and the peak area versus amount of amines was plotted with the linear regression coefficient for histamine, tyramine, putrescine and cadaverine and the correlation coefficient was found greater than 0.99 for each amines (Fig. 4). This proved a definitive linear relationship within amines concentration along with detector response.
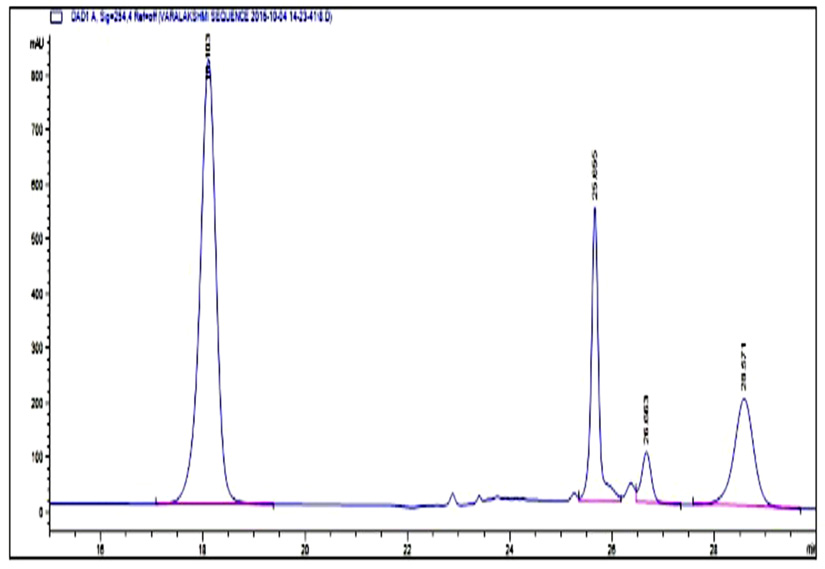
Fig. 5. HPLC Chromatogram of standard BAs Peak retention time at 18.1: Tyramine, 25.6: Putrescine, 26.6: Cadaverine, 28.4: Histamine
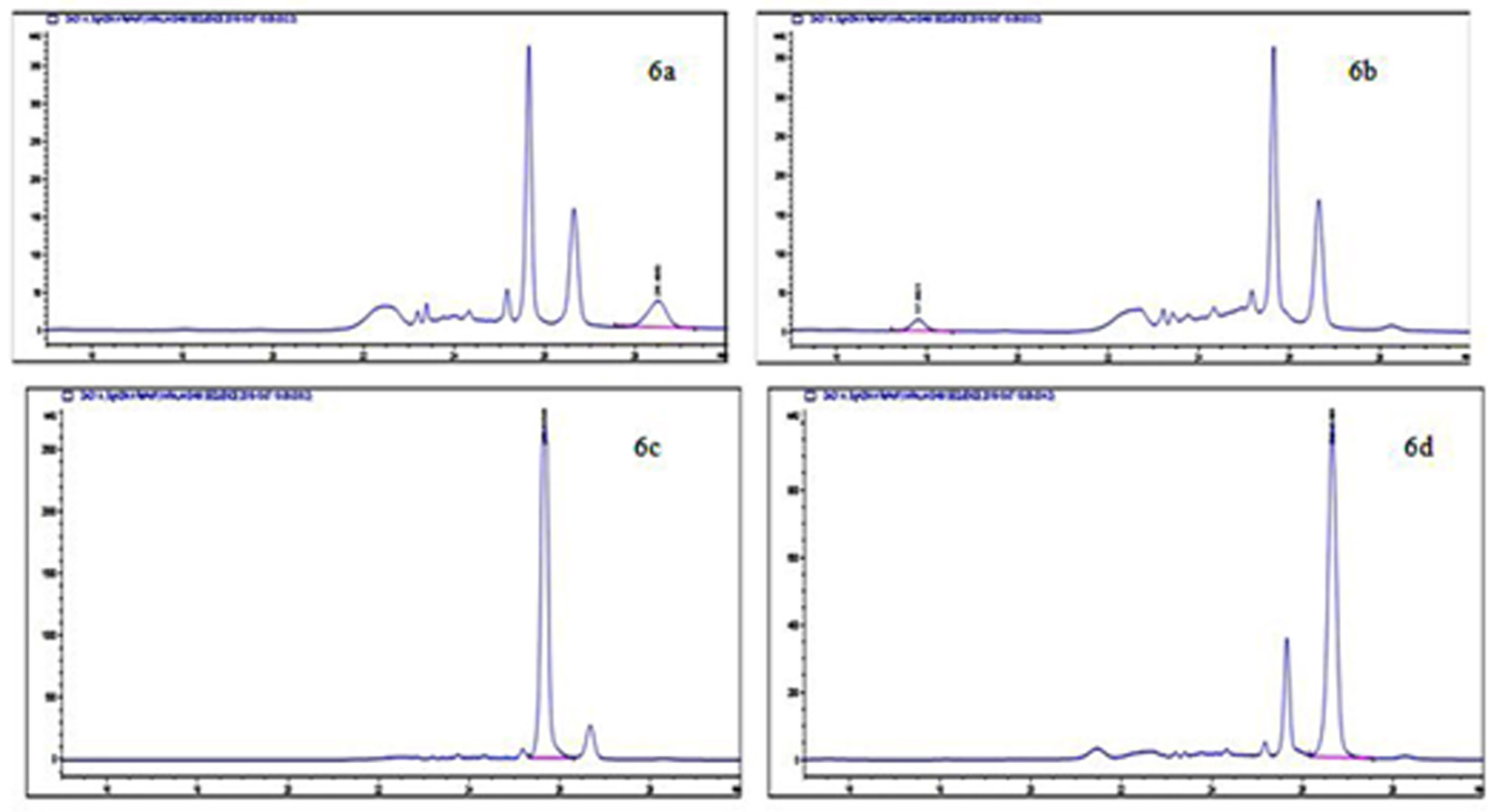
Fig. 6. HPLC Chromatogram of BAs produced by E. cloacae showing a peak of histamine (6a), tyramine (6b), putrescine (6c) and cadaverine (6d)
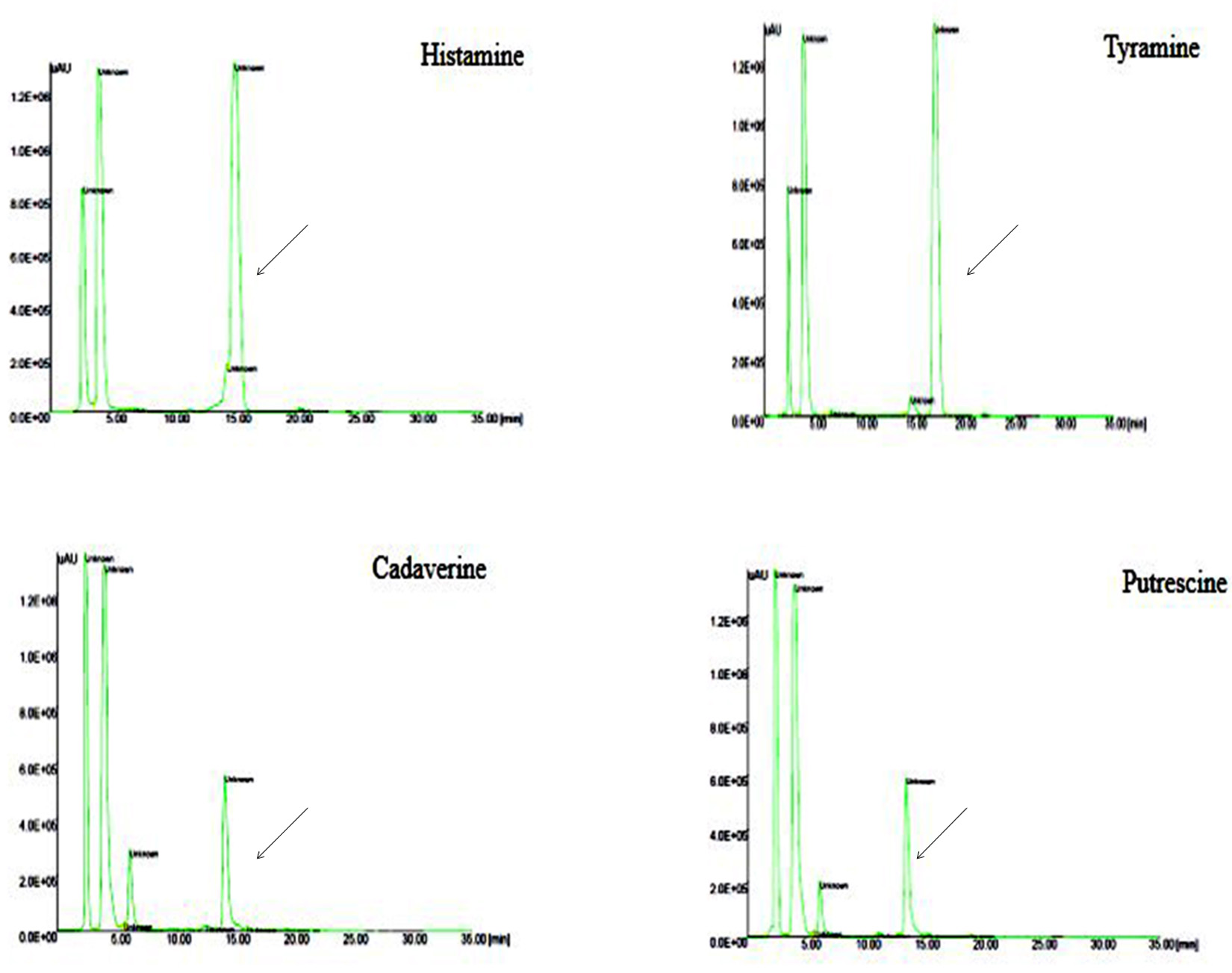
Fig. 7. HPLC Chromatogram of standard BAs Peak retention time at 17.1: Tyramine, 13.4: Putrescine, 14.1: Cadaverine, 14.9: Histamine
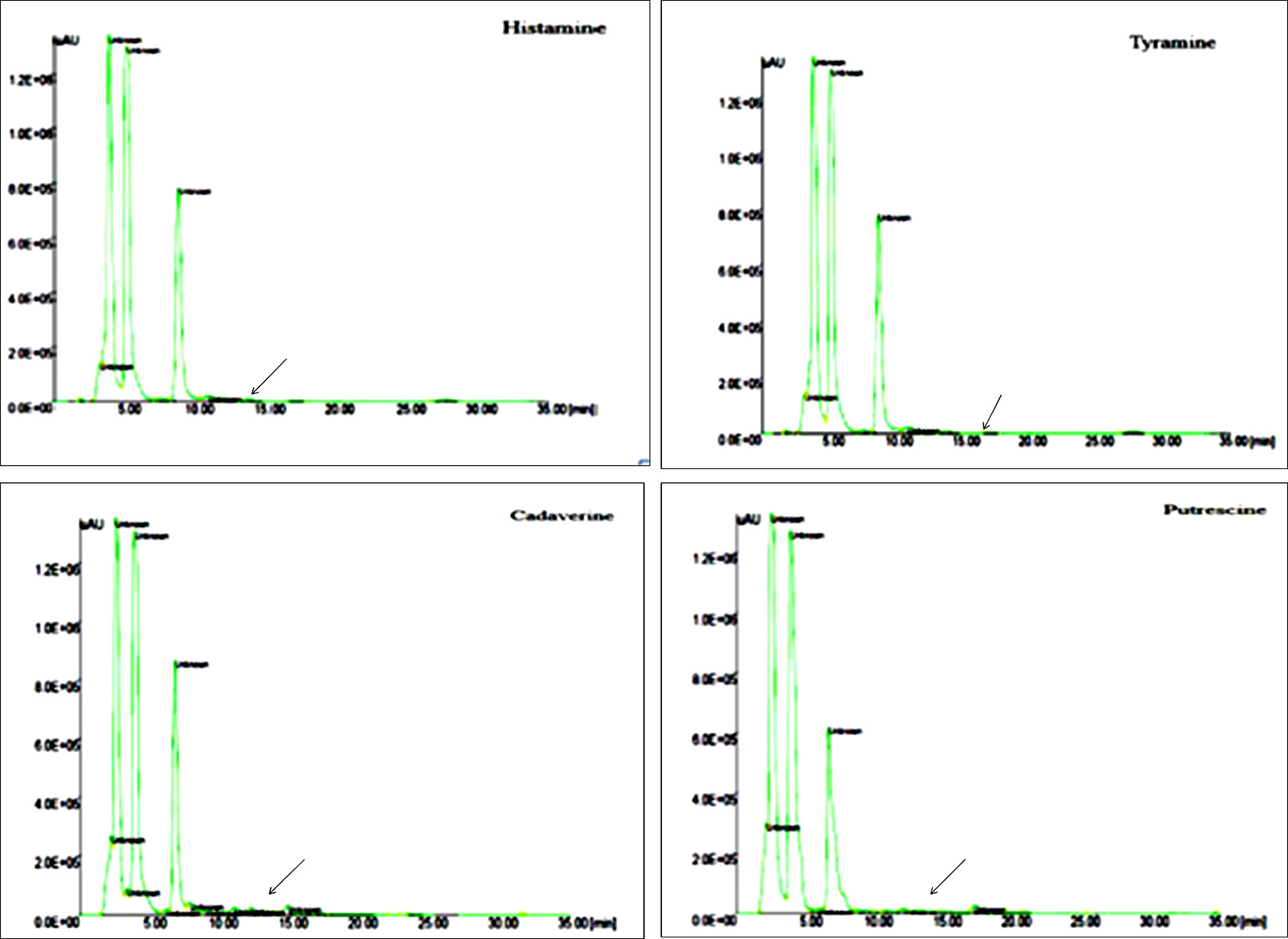
Fig. 8. HPLC Chromatogram of BAs complete degradation of histamine, tyramine and partial degradation of cadaverine and putrescine by L. plantarum.
In the present study, among all BAs producing bacteria, Enterobacteriaceae species namely E. cloacae, E. hormaechei, M. morganii produced the highest amount of putrescine (244, 61, 68 mg/100 gm) followed by cadaverine (154, 208, 15.8 mg/100 gm) and histamine (60.1, 50, 210 mg/100 gm). E. cloacae was producing highest amount of putrescine and cadaverine compared other microorganisms that were BAs positive. The quantity of BAs recorded in the present investigation by E. cloacae were observed to be quite very high compared to the earlier reports of these BAs producing organism recovered from fermented pork sausages (Bover-cid et al. 2001a). Similarly, E. hormaechei in the present investigation recovered from kanji had relatively high level of BAs when compared with the strains of E. hormaechei reported earlier from dairy sources (Coton et al. 2012). As observed from the previous investigation (Bjornsdottir et al. 2009), M. morganii was found to be prolific histamine producing bacteria among Enterobacteriaceae family in our investigation as well.
licheniformis other than Entero-bacteriaceae family recovered from sauerkraut produced putrescine, cadaverine and histamine (58.3, 83.3, 31.7 mg/100 gm) far in excess, than the B. licheniformis recovered from Myeolchi-jeot in the earlier report where the yield were below 4-6 µg/ml (Mah et al. 2003). In the recent report of Moracanina et al. (2015), L. sakei recovered from uzicka sausage produced both tyramine and histamine in the microgram level. Whereas, a strain of the same species recovered from meat pickle in the present study produced only tyramine in milligram level.
In case of degradation study degradation percentage of BAs is quantitatively detected by sensitive HPLC method (detection limit 4 10 ng/mL). where L. plantarum able to degrade 100% of histamine and 100% of tyramine, 28% of putrescine and 67.8% of cadaverine therefore L. plantarum strains were selected as good candidates for further study. L. plantarum from canine saliva shows high percentage of degradation and higher the cell concentration of L. plantarum lower was the BAs content and a positive correlation was observed. Few studies showed L. plantarum isolated from wine degraded histamine, tyramine, putrescine and cadaverine (Garcםa ruiz et al 2011). Recently Xia et al. 2018 reported consortium of L. plantarum and Staphylococcus xylosus were in the ratio of 1:2 significantly reduced histamine (42.6%), tyramine (42.8%) and putrescine (43.0%). L. plantarum from canine found to be best candidate for BAs degrading bacteria as observed in our investigation as well.
Worldwide the fermented products are in high demand because of their nutritive value, but on the other hand, fermented foods are also a potential source of BAs because of microbial contamination during processing or raw materials present in it or on storage BAs might have formed. In the present work E. hormaechei, E. cloacae from kanji, B. licheniformis from sauerkraut, M. morganii from fish pickle and L. sakei from meat pickle were proved to be the potential producers of BAs. Different BAs studied in the present investigation ranged from 3 to 2440 mg/100 gm of sample therefore screening of starter cultures should be made mandatory in order to provide the safe fermented products. Alimentation of better manufacturing methods, advancement of production technology and development of the hygienic condition of raw materials used for manufacturing various types of fermented products are required in order to provide the safe consumption of these products with respect to their BAs content and also finding of this research shows that within the L. plantarum of canine saliva able to degrade histamine, tyramine, cadaverine and putrescine. It may also be concluded that the amines degradation activity was rather strain specific than species specific. The amine oxidase activity as well as the safety of these bacteria can be considered as the criteria for the bacteria used to reduce amine accumulation. This may lead to a favorable contribution to public health, especially in fermented foods in the diet.
The authors are thankful to the Director, Defence Food Research Laboratory, Mysore for providing the lab facility to carry out research work.
The authors declare no conflict of interest.
- Bה M, Moellecken U, Kצnig H, Claus H. The Potential of the Yeast Debaryomyces hansenii H525 to Degrade Biogenic Amines in Food. Microorganisms, 2015; 3: 839-850.
- Bjornsdottir K, Bolton G.E, McClellan-Green P.D, Jaykus L.A, Green D.P. Detection of gram-negative histamine-producing bacteria in fish: a comparative study. Journal of Food Protection, 2009; 72(9): 1987–1991.
- Bover-Cid S, Hugas M, Izquierdo-Pulido M, Vidal-Carou M.C. Amino acid-decarboxylase activity of bacteria isolated from fermented pork sausages. International Journal of Food Microbiology, 2001; 66(2001): 185–189.
- Chen H.C, Huang Y.R, Hsu H.H, Lin C. Saint, Chen W.C, Lin C.M, Tsai Y.H. Determination of histamine and biogenic amines in fish cubes (Tetrapturus angustirostris) implicated in a food-borne poisoning. Food Control, 2010; 21: 13–18.
- Coton M, Delbיs-Paus C, Irlinger F, Desmasures N, Le Fleche A, Stahl V, Montel M.C, Coton E, Diversity and assessment of potential risk factors of Gram-negative isolates associated with French cheeses. Food Microbiology, 2012; 29: 88–98.
- FDA. Bacteriological analytical manual. Arlington, VA: AOAC International 1992. https://www.fda.gov/food/foodscienceresearch/laboratorymethods/ucmHYPERLINK “https://www.fda.gov/food/foodscienceresearch/laboratorymethods/ucm2006949.htm”2006 949HYPERLINK“https://www.fda.gov/food/foodscienceresearch/laboratorymethods/ucm2006949.htm”.htm
- Garcםa-ruiz A, Gonzבlez-rompinelli EM, Bartolomי B, Moreno-arribas MV. Potential of wine-associated lactic acid bacteria to degrade biogenic amines. International Journal of Food Microbiology, 2011; 148(2): 115-120.
- Gardini F, “ײzogul Y, Suzzi G, Tabanelli G, “ײzogul F. Technological factors affecting biogenic amine content in foods: A review. Frontiers in Microbiology, 2016; 7: 1–18.
- Hugas M, Garriga M, Aymerich T, Monfort J.M. Biochemical characterization of lactobacilli from dry fermented sausages. International Journal of Food Microbiology, 1993; 18: 107-113.
- Jeya Shakila R, Vasundhara T.S, Kumudavally K. V. A comparison of the TLC-densitometry and HPLC method for the determination of biogenic amines in fish and fishery products. Food Chemistry, 2001; 75: 255–259.
- Loukou Z and Zotou A. Determination of biogenic amines as dansyl derivatives in alcoholic beverages by high-performance liquid chroma-tography with fluorimetric detection and charac-terization of the dansylated amines by liquid chromatography atmospheric pressure chemical ionization mass spectrometry. Journal of Chromatography A., 2003; 996: 103–113.
- Luten, J. B, Bouquet W, Seuren L. A. J, Burggraaf M. M, Riekwel-Booy G, Durand. Biogenic amines in fishery products: standardization methods within EC. In H. H. Huss (Ed.), Quality assurance in the fish Amsterdam. Elsevier Science Publishers, 1992: 427-439.
- Mah J.H, Ahn J.B, Park J.H, Sung H.C, Hwang H.J. Characterization of biogenic amine-producing microorganisms isolated from Myeolchi-jeot, Korean salted and fermented anchovy. Journal of Microbiology and Biotechnology, 2003; 13: 692–699.
- Marcobal A, De Las Rivas B, Muסoz R. Methods for the detection of bacteria producing biogenic amines on foods: A survey. Journal f r Verbraucherschutz und Lebensmittelsicherheit, 2006; 1: 187–196.
- Mo Dugo G, Vilasi F, La Torre G.L, Pellicanע T.M. Reverse phase HPLC/DAD determination of biogenic amines as dansyl derivatives in experimental red wines. Food Chemistry, 2006; 95: 672–676.
- Moracanina S.V, Stefanovic S, Radicevic T, Borovic B, Djukic D. Production of biogenic amines by lactic acid bacteria isolated from Uzicka sausages. Procedia Food Science, 2015; 5: 308–311.
- Moreno-Arribas M.V, Polo M.C, Jorganes F, Muסoz R. Screening of biogenic amine production by lactic acid bacteria isolated from grape must and wine. International Journal of Food Microbiology, 2003; 84: 117–123.
- Naila A, Flint S, Fletcher G, Bremer P, Meerdink G. Control of Biogenic Amines in Food — Existing and Emerging Approaches. Journal of Food Science, 2010;75(7): 139-150.
- ײnal A. A review: Current analytical methods for the determination of biogenic amines in foods. Food Chemistry, 2007; 103: 1475–1486.
- Ranjekar A, Mahagaonkar R, Mujumdar S. Original Research Article Multi-drug Resistance of Micro-Organisms Isolated from Dog Skin and Saliva. International Journal of Current Microbiology and Applied Sciences, 2015; 2: 317-329.
- Sambrook J, Fritsch E. F, Maniatis T. Molecular Cloning: A Laboratory Manual, second ed. Cold Spring Harbor Laboratory Press, New York 1989.
- Taylor P, Prester L, Chemistry, Analysi, Control, Exposure & Risk Assessment Biogenic amines in fish,fish products and shellfish/ : a review. Food Additives & Contaminants, 2011; 28: 1547-1560.
- ten Brink B, Damink, C, Joosten H.M.L.J, Huis in ’t Veld J.H.J. Occurrence and formation of biologically active amines in foods. International Journal of Food Microbiology, 1990; 11: 73–84.
- Vidal-Carou M.C, Izquierdo-Pulido M.L, Martםn-Morro M.C, Marinי-Font. Histamine and tyramine in meat products: Relationship with meat spoilage. Food Chemistry, 1990; 37: 239–249.
- Weisburg, W.G, Barns S.M, Pelletier D.A, Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology 1991; 173: 697-703.
- Xia X, Luo Y, Zhang Q, Huang Y, Zhang B. Mixed Starter Culture Regulate Biogenic Amines Formation via Decarboxylation and Trans-amination during Chinese Rice Wine Fermen-tation. Food and Beverage Chemistry Biochemistry, 2018: 1-32.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.