ISSN: 0973-7510
E-ISSN: 2581-690X
Bacterial strain identified as Klebsiella pneumoniae SK1, based on 16S rDNA sequence study, was isolated from a textile mill discharge point at Dada Nagar industrial area in Kanpur, India in media containing 100 µg/ml each of disulfonated azo dye Acid Red 249 (AR) and hexavalent chromium [Cr(VI)]. SK1 is efficient in the concomitant decolorization and reduction of 100 µg/ml toxic AR and Cr(VI) respectively in 48 h under microaerophilic conditions. The efficacy of AR decolorization was found to be preeminent in static conditions, 37°C, 2-4% salinity and 7-9 pH range. 99% decolorization for initial AR concentration of 100 µg/ml in 48 h, and 91% for 250 µg/ml and 77% for 500 µg/ml was recorded in 72 h respectively in the presence of 100 µg/ml Cr(VI). Significant reductive changes in spectroscopic absorption spectra were observed for SK1 treated AR+Cr(VI) amended media with respect to controls. FITR spectroscopy was used to ascertain the breakage of the azo dye bond and the formation of biodegradative metabolites. Additionally, SK1 was found to be positive for indole acetic acid, ammonia, phosphate and potassium solubilization and biofilm formation. In plant bioassay, in vitro SK1 treated AR+Cr(VI) TSB media was used to treat Triticum aestivum in the soil environment. In comparison to untreated control, plants treated with bioremediation media shows increased percent germination, root and shoot length with a complete reversal of phytotoxic effects of the AR+Cr(VI) treated plants. In this study, environmental K. pneumoniae SK1 shows concomitant azo dye and Cr(VI) remediation with plant growth-promoting activity.
Acid red azo dye and Cr(VI) bioremediation, Plant growth promotion, Klebsiella pneumoniae, Phytotoxicity
Textile and leather industry are a prolific source of environmental pollutants such as heavy metal chromates and its complexes as well as synthetic azo dyes1. Concurrent existence of hexavalent chromium Cr(VI) and azo dye in water bodies and soil resulting from tannery and textile industries have detrimental impact on all living forms when released into the environment without any treatment2. Cr(VI) is a priority pollutant as per United States Environment Protection Agency with carcinogenic and mutagenic properties due to its high oxidizing nature and water solubility3. Xenobiotics such as sulfonated azo dyes are a cause of serious public health and environmental hazard due to their recalcitrance and physico-chemical properties of causing color and odor contamination of natural resources2. The compounds contain difficult to degrade azo bonds (-N=N-) as well disulfonated aromatic (SO43-)4. Both disulfonated azo dyes and chromate possess toxic properties and their co-existence in the industrial spent released into the treatment systems or directly into the surrounding environmental leads to further synergistic toxicity effects to all life forms. A bioremediation strategy that targets the simultaneously azo dye decolorization as well as Cr(VI) reduction is a viable, effective, economical and ecofriendly alternative. Textile effluent sludge are a rich source of inorganic and organic nutrients providing a natural environment for the isolation of indigenous Cr(VI) and azo dye resistant bacteria5. Bioremediation of azo dyes and Cr is reported previously for either Cr(VI) or azo dye alone by several microbial systems, but only a few reports describe the concomitant decolorization of azo dye and Cr(VI) reduction6,7. Redox enzymatic steps are involved in the azo bond cleavage as well as Cr(VI) reduction, which make bacteria capable of carrying out both reactions important natural bioresources. Most remediation strains use reducing enzymes (azoreductases, laccases, peroxidases, superoxide dismutase, catalase) under aerobic/microaerophilic conditions to break down azo dyes into colorless aromatics amines and subsequently into non-toxic amines8. Anaerobic degradation using oxygen sensitive azoreductase has also reported to be a more efficient process9. Similarly, Cr(VI) reduction has been reported to occur in aerobic or anaerobic or under both conditions in different microbial systems involving one or more metabolic pathways10.
The objective of the present report, was to screen for azo dye decolorizing and Cr(VI) reducing bacteria from textile spent. Herein, is a first report of a textile spent isolate Klebsiella pneumoniae SK1 with the ability to concomitantly decolorize disulfonated azo dye Acid Red (AR), reduce Cr(VI) and show plant beneficial properties (PBP). The effect of AR and Cr(VI) concentrations on the remediation ability of either was further evaluated. The strain was characterized by the 16S rRNA gene sequence analysis, and its performance in azo dye decolorization in variable environmental parameter, Cr(VI) bio-reduction and plant growth-promoting activity in simultaneous manner under soil condition reported. Functional phytotoxicity characterization of the remediated AR and Cr(VI) solution using Triticum aestivum (wheat) is described.
Isolation, characterization and identification
Microbial enrichment was carried out using industrial textile dye sludge [1.0 g, collected from Dada Nagar industrial area Kanpur, UP, India (26.4534° N, 80.2941° E)] in 100 ml tryptone soya broth (TSB) amended with 100 µg/ml of both Acid Red 249 (AR) and K2Cr2O7 [Cr(VI)]. Media and chemicals used in the study were purchased from HiMedia, India and Merck India. Further, bacterial isolates were sub culturing on tryptone soya agar (TSA) plate amended with 100 µg/ml of both AR and Cr(VI). Colonies with potential decolorizing zone were isolated and purified for further experiment. Identification of bacterial isolate was carried out by 16S rDNA sequence analysis. Genomic DNA was amplified by PCR using consensus primers and sequenced (Agrigenome Labs Pvt Ltd., Kochi, Kerala, India). The DNA sequence obtained was analyzed at National Center for Biotechnology Information server using BLAST tool and corresponding sequences downloaded (www.ncbi.nih.nlm.in). Phylogenetic tree was constructed using neighbor joining program in MEGA X11.
Minimum Inhibitory Concentration (MIC) and Cr(VI) effect on AR decolorization
To determine the MIC, the isolate was sub cultured in increasing concentrations of (100, 250, 500, 750, 1000, 1250 and 1500 µg/ml) AR and Cr(VI) in TSA separately until day 5 post incubation at 37°C12. The effect of varying Cr(VI) concentrations 100, 250 and 500 µg/ml on efficacy of decolorization was evaluated on TSA plate containing AR at different concentration of 100, 250 and 500 µg/ml on the basis of individual MIC.
Decolorization Assay
TSB media amended with different concentration of 100, 250 and 500 µg/ml of AR with initial concentration of 100 µg/ml Cr(VI) was inoculated with 24 h log phase culture (10% v/v) and incubated at 37°C for 48 hours. The samples from culture were centrifuge at 10000 rpm for 10 minutes. Supernatant from centrifuge sample was analyzed using spectrophotometer (MULTISKAN EX, Thermo Fisher Scientific, USA) at 520 nm. The decolorization percent of AR by isolates using spectrophotometer were described as per following equation13:
Decolorization % = ((Initial absorbance-Final absorbance) X 100) / (Initial absorbance)
…(1)
Optimization of the AR decolorization parameters
Effect of varying parameters such as pH, temperature, oxygen and salinity on SK1 mediated decolorization and growth kinetic was analyzed in TSB media containing 100 µg/ml AR+Cr(VI) as described previously14. The effect of oxygen concentration on decolorization was analyzed by incubating the cultures separately in static and shaken (200 rpm) for 48 respectively. Effect of pH on AR decolorization was assessed in TSB media adjusted to pH 5, 7 and 9. The effect of various concentrations of NaCl ranging from 2%, 4% and 6% was also investigated in TSB medium under the same operational conditions. For optimization of temperature, isolate was grown in varying temperature 25, 37 and 45°C respectively. Percent decolorization and growth were measured for each experiment under environmental parameter daily.
Biodegradation analysis
Spectrophotometric analysis was carried out from 400-700 nm to observe the change in absorption spectra of decolorized AR compare to non-inoculated AR media, against base line defines using AR free media as control 15. The complete degradation of AR was also confirmed by ATR FTIR spectroscopic analysis (Tensor 27 Bruker Germany, Center for Environmental Science and Engineering, IIT Kanpur) 16. Treated and untreated media amended with 100 µg/ml each AR+Cr(VI) were centrifuged at 10,000 rpm for 10 min (GeNei Centrifuge model SLM). Supernatant samples were scanned at 4000-500 cm-1 using 16 scans and FTIR spectra acquired as absorbance values. Spectra obtained were analyzed using the Origin software.
Concomitant bioremediation of Acid Red and Cr(VI)
To evaluate the ability of the isolate to detoxify simultaneously AR and Cr(VI), the isolate was cultured in TSB media amended with 100 µg/ml each AR+Cr(VI) and incubated at 37°C in microaerophilic condition17. Aliquots were removed by different time intervals and decolorization percent measured using spectrophotometry and Cr(VI) levels determined using the 1,5 diphenyl carbazide method 18. Briefly for Cr(VI) bioreduction, sample aliquots were acidified using 6N H2SO4 followed by addition of 25% (w/v) 1, 5 diphenyl carbazide in acetone and Cr(VI) presence detected by spectrophotometer (Thermo Scientific, Spectronics) at 540 nm. Controls included media with and without Cr(VI). In concomitant samples, Cr(VI) reduction was measured only post complete AR decolorization when there was no color interference.
Plant growth-promoting product (PGPP) and Biofilm formation
All the PGPP and biofilm formation assays were performed in in vitro condition. Phosphate solubilization was analyzed by measuring the clearance zone on Pikovskaya’s agar medium and potassium solubilization was checked on Aleksandrow agar medium19. Indole acetic acid (IAA) production was tested using tryptone broth containing 0.1% tryptophan. The medium was incubated with 10% log culture (107 cell/ml) for 2 days at 37°C with shaking. After centrifugation at 10,000 g for 10 minutes, 1 ml of supernatant was mixed with 2 ml of Salkowasky’s reagent and the optical density was measured at 550 nm20. Ammonia production ability of SK1 was analyzed by using peptone water at 37°C for 2 days. After incubation, 1 ml of Nessler’s reagent was added into each tube and the development of yellow color indicates ammonia production and measured absorbance at 450 nm21.
Biofilm formation was assayed with brief modifications22. 500 µl log phase cultures (5×106) in micro centrifuge tubes were incubated at 37°C for 24 h. Post incubation, culture media was decanted and biofilm associated with microcentrifuge tube stained with crystal violet (1% w/v) for 20 min. After washing excess crystal violet, dimethyl sulphoxide (DMSO) was added to mobilize crystal violet stained bacteria. The biofilm and associated planktonic cell absorbance at 620 nm were measured spectrophotometrically in a microplate reader (MULTISKAN EX, Thermo Fisher Scientific, USA) and the Biofilm Index (BI) determined as
BI = (Absorbance of biofilm at 620 nm ) /(Absorbance of associated planktonic cells at 620 nm)
…(2)
Phytotoxicity Assay
Phytotoxicity effect of AR and degraded products was assayed on seed germination and plant growth of Triticum aestivum23. Media containing 100 µg/ml AR and Cr(VI) each was treated with SK1 isolate under optimal conditions for 48 h. Un-inoculated media with 100 µg/ml AR and Cr(VI) was used as control. Media was centrifuged at 6000 rpm for 10 minutes and 1:1 diluted with distilled water. Pot experiments containing sterile soil at room temperature were planted with 10 seeds and 5 ml of each treatment used to irrigate the seeds on alternate days. Suitable controls were made with sterile water. Percent seed germination and length of shoot and root were recorded after 12 days.
Statistical analysis
Results are presented as either mean ± SEM (standard error of mean) or SD (standard deviation) using statistical t test (p ≤ 0.05) for normally distributed data and median for non- normal distribution data. The multiple comparisons among treatments were made using ANOVA with the Kruskal–Wallis test.
Microbial Enrichment, Characterization and Identification
each for 48 h at 37°C, one bacterial isolate SK1, was found to be a fast grower and showed AR decolorization in primary screening test and therefore was selected for further microbial and biochemical characterization. SK1 was characterized to be oxidase positive, catalase positive, capsule forming, lactose non-fermenting, endospore non-forming and Gram-negative small rod. Identification of bacterial isolate was carried out by 16S rDNA sequence analysis. The amplified sequence showed 99% similarity to Klebsiella pneumoniae (Gen Bank Accession Number KY417867). On the basis of 16S rRNA gene sequence analysis, the isolate (SK1) was identified as Klebsiella pneumoniae SK1 and submitted with NCBI GenBank accession No. MK902672.1. The phylogenetic tree was constructed using the neighbor joining program in MEGA X analyze pair wise alignment between the related sequences for each Klebsiella pneumoniae strains (Fig. 1). Previously, Klebsiella species (Bz4 and Rz7) have been described as decolorization of different dyes such as bengal rose, brilliant green, erythrosine, crystal violet and azo dye Evans blue23.
Fig. 1. Phylogenetic tree derived from 16S rRNA gene sequence of Klebsiella pneumoniae SK1 (MK902672.1), using neighbor joining program in MEGA X
Minimum Inhibitory Concentration of AR and Cr(VI)
The effects of separate treatments of increasing concentrations of 100-1500 µg/ml AR and Cr(VI) respectively were studied on SK1 growth and AR decolorization ability. Isolate SK1 was found to show growth up to 1250 µg/ml AR, although decolorization was found only up to 500 µg/ml AR following 48 h incubation. MIC of only Cr(VI) was observed to be 1000 µg/ml in 48 h incubation on TSA plate (Table 1A).
Based on individual Cr(VI) and AR MIC and decolorization data, concomitant effects of varying concentrations on AR and Cr(VI) on SK1 growth and decolorization ability were studied. Decolorization ability of isolate at 100 µg/ml AR was found to be unaffected by the presence of 100 µg/ml Cr(VI), while further increasing Cr(VI) concentration severely inhibit AR decolorization (Table 1B). The inhibitory effect of 100 µg/ml Cr(VI) was found on 500 µg/ml AR, 250 µg/ml Cr(VI) on 250 µg/ml AR and 500 µg/ml Cr(VI) on 100 µg/ml AR decolorization. Interestingly, no effect on the growth of the isolate was recorded in some of the combinations, which still negatively affected azo dye decolorization. Hence, it is likely that stress caused by increased concentrations of AR+Cr(VI) causes shut down of enzymes systems involved in the reduction of the xenobiotics. A previous study also reported that increasing the concentration of Cr(VI) from 0 to 150 µg/ml resulted in the decreased efficiency of Lactobacillus paracase to decolorize acid black with initial concentration of 100 µg/ml17. Cr(VI) concentration above 30 µg/ml also severely inhibited the azo dye bioremoval by Tinea versicolor24.
Table (1A):
MIC of AR and Cr(VI) individually on growth and ZOD in TSA media.
Concen. (µg/ml) |
AR MIC |
AR ZOD |
Cr(VI) MIC |
|---|---|---|---|
100 |
+++ |
+++ |
+++ |
250 |
+++ |
++ |
+++ |
500 |
+++ |
+ |
++ |
750 |
+++ |
– |
+ |
1000 |
++ |
– |
– |
1250 |
+ |
– |
– |
1500 |
– |
– |
– |
Table (1B):
Effect of varying concentration concomitant Cr(VI) and AR on growth and ZOD in TSA media.
| Concen. Cr (VI)+AR (µg/ml) |
Growth | ZOD | |
|---|---|---|---|
| 24 h | 24 h | 48 h | |
| 100 + 100 | +++ | ++ | +++ |
| 100 + 250 | +++ | – | ++ |
| 100 + 500 | ++ | – | – |
| 250 + 100 | +++ | – | + |
| 250 + 250 | ++ | – | – |
| 250 + 500 | ++ | – | – |
| 500 + 100 | ++ | – | – |
| 500 + 250 | ++ | – | – |
| 500 + 500 | + | – | – |
+++: Heavy growth and zone of decolorization,
++: Lesser growth and zone of decolorization
+: Very less growth and zone of decolorization
-: no decolorization even after 96 h incubation
ZOD: Zone of decolorization
Effect of Oxygen, pH, Temperature and Salinity on Acid Red decolorization
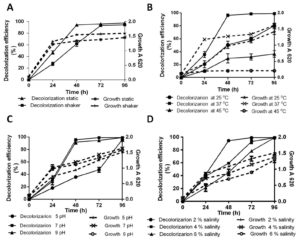
In the natural environment, strongly fluctuating abiotic parameters due to release of toxic waste from industrial effluents, detrimentally affect bioremediation ability. The effect of various parameters such as oxygen requirement, temperature, pH and salinity on AR decolorization in the presence of 100 µg/ml Cr(VI) was assayed to characterization of optimum condition. Fig. 2A depicts color removal efficiency and growth kinetic through 96 h of incubation period under static and continuous shaking condition. SK1 shows similar growth in static and shaker at the end of 24 h. However, 14%, 12% and 8% increment in growth was obseved in shaker condition comparared to static at 48 h, 72 h and 96 h respecively. During static conditions, SK1 was capable of color removal up to 94% within 48 h while in the same time duration, 62% decolorization was observed under shaker conditions. Complete decolorization of AR was also observed under shaker conditions in 72 h incubation period. SK1 is therefore capable of AR decolorization in both aerobic and microaerophilic conditions with the latter being most efficient. In a previous study, facultative bacteria E. coli and Pseudomonas sp failed to decolorize DB 38 and CR under aerobic conditions but when the same cultures were incubated under anaerobic and microaerophilic conditions significant decolorization was achieved9. Bacterial decolorization of azo dyes occurs due to enzymatic activity based electron transfer to azo group from NADH under anaerobic condition25. The second stage decolorization of mono-, di- and tri- sulfonated azo dyes in 24 h followed by aerobic degradation of 4 amino sulfobenzoic acid has been reported for K. pneumoniae8.
Fig. 2. Percentage decolorization of 100 µg/ml AR with 100 µg/ml Cr(VI) and growth kinetic by SK1; static and shaken (A), variable temperature 25°C, 37°C and 45 ºC (B), pH 5, 7 and 9 (C) and Salinity 2%, 4%, 6% (D)
Effect of temperature on 100 µg/ml AR decolorization is shown in Fig. 2B. It can be inferred that maximum AR decolorization up to 90% occurred at 37°C during 48 h of incubation whereas in the same time period, significant decrease in both decolorization (27% and 29%) and growth (23% and 83%) was recorded at 25°C and 45°C respectively. Decolorization efficiency of SK1 was 95% at pH 7 and 92% at pH 9 during 48 h of incubation with similar growth kinetics (Fig. 2C). SK1 showed significantly lower decolorization and growth rates at pH 5 during similar incubation. In this study, temperature was observed to have a strong effect on decolorization efficeincy of SK1 in comparison to the other parameters. This may be the consequence of temperature variability affecting the efficiency of enzyme mediated AR decolorization. Efficiency of Bacillus subtilis azoreductase in decolorizing four azo dyes (methyl red, congo red, orange G and azobenzene) is influenced by variations in both temperature and pH26. Decline in decolorization efficiency is normally associated with denaturation of azoreductase and loss of cell viability at higher temperature27. Acinetobacter radioresistens mediated degradation and decolorization of Acid Red decreased from 95% in pH 7 to approximately 47% at pH 5 and pH 813. Pseudomonas putida was reported to show 82% decolorization in media contain 100 µg/ml orange II dye at optimum pH 8, which significantly declined to 30% and 44% at pH 7 and 9 respectively28. Most textile and tannery effluents are rich in brine and so the effect of saline stress was checked on dye decolorization ability of SK1. AR decolorization of 94% was recorded at 2% salinity within 48 h and complete decolorization at a wide salinity range (2-6%) within 96 h even as decreased growth kinetics were recorded with increase in salinity concentration (Fig. 2D). More than 90% decolorization efficiency of a moderately halotolerant bacterium, Bacillus firmus in synthetic saline wastewater medium containing 100 µg/ml of Polar red B (azo dye) in a wide range of NaCl concentration 0–6% (w/v) under static condition within 24 h is reported29. In a study with halophilic bacterial isolate Shewanella aquimarina, 14.5% Acid Red 27 was found to be decolorized in media containing 10% salinity30. In our study, SK1 was also found to be halotolerant and capable of AR decolorization over wide (2-6%) salinity range.
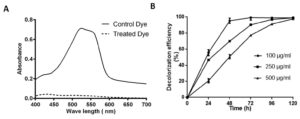
Fig. 3. Absorption spectra of control and SK1 treated 100 µg/ml Acid Red (AR)+Cr(VI) after 48 hours incubation (A), Decolorization percent of AR at different concentration 100, 250 and 500 µg/ml with initial 100 µg/ml Cr(VI) (B).
Decolorization assay of Acid Red
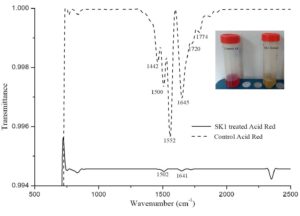
Isolate SK1 was assayed for its ability to decolorize AR under static conditions, pH 7 and temperature 37°C by measuring the absorbance spectra of samples at different time points. Fig. 3A shows, significantly reduced peak at 525 nm in treated sample compared to control. Fig. 3B shows the kinetics of decolorization percentage of AR as a measure of increasing concentration. Decolorization for 100, 250 and 500 µg/ml AR was found to be 95%, 70% and 50% respectively following 48 h of incubation. AR biodegradation by the isolate was confirmed by scanning the control and decolorized samples using ATR FTIR spectroscopy. Fig. 4 shows FTIR analysis of control and decolorized samples, which demonstrates significant differences and shifts in specific peaks of AR in 1000–2000 cm−1 spectrum range. FTIR spectrum of control dye AR showed peaks at 1552, 1500 and 1442 cm-1 which represent the presence of N=N stretching, N-H deformation, C-H deformation respectively. Break down of azo bond was confirmed by the absence of spectral peak at 1552 cm-1. The presence of peak around 2353 cm-1 and 1600 cm1 represent functonal group C-O and amide group respectively in SK1 treated sample which indicate breakdown products of AR. In previous study, peak of 1519 and 1608 cm−1 were also suggested presence of azo bond (N=N) in disperse red 7331,32. Our study also align with FTIR analysis of Acid Red 1 degradation by Stenotrophomonas sp. wherein decrease in peak at 1597 cm-1 indicated azo group breakdown33. Azobacter vinelandii treated Disperse Red 73 amended media showed absence of azo bond (1519 cm-1) in the biodegraded products31.
Fig. 4. FTIR profile of Control 100 µg/ml Acid Red (AR) with Cr(VI) and its decolorized products obtained after 48 hours incubation with SK1
Concomitant quantification of AR decolorization and Cr(VI) reduction
Individual and concomitant kinetics of AR decolorization and Cr(VI) reduction were assayed as described previously in materials and methods under optimized conditions. Table 2 data reveals that in 48 h, in samples wherein individual Cr(V) reduction and AR decolorization was measured, 99% Cr(VI) reduction and 95% AR decolorization was recorded. In concomitantly treated samples, reduction of Cr(VI) and AR decolorization was found to be 99% and 91% respectively at 48 h. Furthermore, AR decolorization was found to 49% and 46% in absence and presence of Cr(VI) respectively at 24 h. Cr(VI) couldn’t be assayed at 0 and 24 h due to interference of color and it is possible that the slight increase in absorbance may be attributed to color due to presence of chromium byproducts. SK1 is therefore capable of concomitant remediation of 100 µg/ml AR+Cr(VI) in 48 h microaerobically.
Table (2):
Concomitant AR decolorization and Cr(VI) reduction by K. pneumoniae SK1 cultured in microaerophilic condition within 48 h of incubation.
| Treatment | Initial Concentration (100 µg/ml) |
Time of Incubation | ||||
|---|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | ||||
| D (OD550) | D (OD550) | Reduction (%) | D (OD550) | Reduction (%) |
||
| Individually | Cr (VI) | 0.1±0.001 | 0.06±0.007 | 99% | 0.06±0.005 | 99% |
| AR | 1.11±0.03 | 0.61±0.009 | 49% | 0.15±0.005 | 95% | |
| Concomitant | Cr (VI) + | not determined | not determined |
not determined |
0.11±0.01 | 99% |
| AR | 1.09±0.03 | 0.63±0.05 | 46% | 0.18±0.01 | 91% | |
AR color is measured as Optical Density (OD550) mean ± SD, n = 3
D: Decolorization
As per t test, no biologically statistical difference found between AR decolorization individually treated AR and Cr(VI)+AR treated groups at 24 h and 48 h. Similarly, no difference in Cr(VI) reduction observed in the presence and absence of AR at 48 h.
100% simultaneously removal of 50 µg/ml methyl orange and 50 µg/ml Cr(VI) under the anaerobic condition with 6% salinity in 36 h is reported for Bacillus circulans6. The bacterial strain Lactobacillus paracase isolated from deep sea sediment of the North Atlantic was capable of removing toxic 100 µg/ml Cr(VI) and Acid Black dye simultaneously with efficiency of 58.5 and 51.9% respectively17. Simultaneous detoxification of reactive Orange dye (200 µg/ml) with 58% and chromate reduction (50 µg/ml) 100% activity is associated with bacterial strain Lysinibacillus sp.34. As far as we are aware, this is the first work to show K. pneumoniae strain showing concomitant bioremediation abilities for toxic disulfonated acid red azo dye as well as Cr(VI)
Production of PGPP and Biofilm formation
Many bioremediation microbial isolates can be additionally beneficial in an indirect manner to the process of bioaugmentation when they have the ability to produce environment friendly secondary metabolites such as plant growth promoting products (PGPP)35. Strains producing PGPP in addition to possessing bioremediation abilities can contribute to the health of fauna of the remediation site as well as contribute via microbe assisted phytoremediation processes. Table 3A indicates that the isolate is positive for indole acetic acid (IAA), ammonia, phosphate and potassium solubilization and biofilm formation. Klebsiella pneumoniae has been previously reported for the production of IAA and phosphate solubilization36. Biofilm formation ability contributes to the increased viability in the polluted environment from heavy metal toxicity as well as increased rhizospheric colonization, improved PGPP and heavy metal sequestration37,38. Bacterial isolates also possess the ability to degrade azo dyes and promote plant growth in a simultaneous manner under controlled conditions39.
Phytotoxicity study
Water bodies containing untreated effluents from dyeing industry represent a great environmental hazard as it is associated with reduced productivity of plant and all associated life forms8. In an experiment, SK1 treated 100 µg/ml AR+Cr(VI), untreated 100 µg/ml Cr(VI)+AR and control sterile water were used to irrigate T. aestivum seeds. Watering of plants with SK1 remediated AR+Cr(VI) solution showed 75% germination while the untreated AR+Cr(VI) showed only 25% germination as compared to 100% seed germination with water alone (Table 3B). Interestingly, SK1 treated plants showed increased shoot and root growth in comparison to untreated as well as control water treated plants, likely due to the presence of PGPP in the spent media. IAA producing K. pneumoniae was found to induced rooting in Cicer arietinum which also contributed to toxicity reversal of azo dye40. In a previous study, seeds of S. vulgare and P. mungo exposed to untreated reactive black 5(RB5) exhibited 50% and 60% germination inhibition respectively as compared to exposure with S. halophila treated RB5 biodegradative metabolites41.
Table (3):
In vitro plant growth promotion by K. pneumoniae SK1 (A) and Phytotoxicity effect of untreated and treated 100 µg/ml AR+Cr(VI) on seed germination and growth of Triticum aestivum plant (B).
| (A) Plant growth promotion* |
(B) Phytotoxicity effect |
||||
|---|---|---|---|---|---|
| Phosphate solubilization zone (cm) |
1.50±0.10 | Parameter | Water | Acid Red Cr(VI) |
SK1 Treated |
| Potassium solubilization zone (cm) |
0.9±0.06 | Seed germination | 100 % | 25 % | 75 % |
| Indole acetic acid production OD530 | 0.6±0.01 | Shoot length# (cm) |
4.15a | 2.70b | 7.50c |
| Ammonia production OD450 | 3.5±0.08 | Root length# (cm) |
4.50a | 1.50b | 6.00ac |
| Biofilm Index OD660 |
3.1±0.10 | ||||
OD: Optical Density
*: Data represent as Mean±SD
#: Data are represented as median.
For the same parameters, values with different letters are significantly different (P < 0.05).
The multiple comparisons among treatments were made using ANOVA with The Kruskal–Wallis test
The present study demonstrates the potential of Klebsiella pneumoniae SK1 for concomitant remediation of disulfonated azo dye Acid Red and Cr(VI). Alkalophilic and halotolerant feature of isolated SK1 is significant for the bioremediation of textile wastewater in alkaline and salinity stress. Biofilm ability of isolate is significant for potential PGPP and better survival in stress conditions. SK1 treated AR spent was found to not only significantly reverse AR and Cr(VI) induced stress in Triticum aestivum, but also promoted plant growth. The isolated bacteria could be very effective for plant growth promotion and reduced phytotoxicity effect which is a major concern in agriculture productivity. This is the first report of alkaliphilic and halotolerant Klebsiella pneumoniae strain capable of concomitant Acid Red and Cr(VI) detoxification with plant growth-promoting properties.
ACKNOWLEDGMENTS
We are grateful for ATR FTIR spectroscopic analysis, Center for Environmental Science and Engineering, IIT Kanpur, India.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
VK and SDK concieved the research. VK performed the experiments. VK and PDU designed the figures and tables. SDK supervised and reviewed the manuscript. All authors read and approved the manuscript.
FUNDING
This work was supported financially by the University Grants Commission, New Delhi, India as Seiner Research Fellowship (357996).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Islam M, Mostafa M. Textile dyeing effluents and environment Concerns – A review. J Environ Sci Nat Resour. 2019;11(1-2):131-144.
Crossref - Lellis B, Favaro-Polonio CZ, Pamphile JA, Polonio JC. Effects of textile dyes on health and the environment and bioremediation potential of living organisms. Biotechnol Res Innov. 2019;3(2):275-290.
Crossref - Achmad RT, Budiawan, Auerkari EI. Effects of chromium on human body. Annu Res Rev Biol. 2017;13(2):1-8.
Crossref - Sarkar S, Banerjee A, Halder U, Biswas R, Bandopadhyay R. Degradation of synthetic azo dyes of textile industry: a sustainable approach using microbial enzymes. Water Conserv Sci Eng. 2017;2(4):121-131.
Crossref - Mahmood R, Sharif F, Ali S, Hayyat MU. Enhancing the decolorizing and degradation ability of bacterial consortium isolated from textile effluent affected area and its application on seed germination. Sci World J. 2015;2015:628195.
Crossref - Liu W, Liu C, Liu L, et al. Simultaneous decolorization of sulfonated azo dyes and reduction of hexavalent chromium under high salt condition by a newly isolated salt-tolerant strain Bacillus circulans BWL1061. Ecotoxicol Environ Saf. 2017;141:9-16.
Crossref - Mahmood S, Khalid A, Mahmood T, Arshad M, Ahmad R. Potential of newly isolated bacterial strains for simultaneous removal of hexavalent chromium and reactive black-5 azo dye from tannery effluent. J Chem Technol Biotechnol. 2013;88(8):1506-1513.
Crossref - Dixit S, Garg S. Biodegradation of environmentally hazardous azo dyes and aromatic amines using Klebsiella pneumoniae. J Environ Eng. 2018;144(6):04018035.
Crossref - Isik M, Sponza DT. Effect of oxygen on decolorization of azo dyes by Escherichia coli and Pseudomonas sp. and fate of aromatic amines. Process Biochem. 2003;38:1183-1192.
Crossref - Thatoi H, Das S, Mishra J, Rath BP, Das N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: A review. J Environ Manage. 2014;146:383-399.
Crossref - Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547-1549.
Crossref - Rahul, Kumar V, Kaistha SD. Hexavalent chromium bioreduction and its functional characterization using Vigna radiata. Res J Environ Sci. 2018;12(4):192-197.
Crossref - Ramya M, Iyappan S, Manju A, Jiffe JS. Biodegradation and decolorization of Acid Red by Acinetobacter radioresistens. J Bioremediation Biodegrad. 2010;1:105.
Crossref - Kang C, Wu P, Li Y, Ruan B, Zhu N, Dang Z. Estimates of heavy metal tolerance and chromium(VI) reducing ability of pseudomonas aeruginosa CCTCC AB93066: Chromium(VI) toxicity and environmental parameters optimization. World J Microbiol Biotechnol. 2014;30(10):2733-2746.
Crossref - Singh AL, Chaudhary S, Kayastha AM, Yadav A. Decolorization and degradation of textile effluent with the help of Enterobacter asburiae. Indian J Biotechnol. 2015;14(1):101-106.
- Desai C, Jain K, Patel B, Madamwar D. Efficacy of bacterial consortium-AIE2 for contemporaneous Cr(VI) and azo dye bioremediation in batch and continuous bioreactor systems, monitoring steady-state bacterial dynamics using qPCR assays. Biodegradation. 2009;20(6):813.
Crossref - Huang G, Wang W, Liu G. Simultaneous chromate reduction and azo dye decolourization by Lactobacillus paracase CL1107 isolated from deep sea sediment. J Environ Manage. 2015;157:297-302.
Crossref - Sagar S, Dwivedi A, Yadav S, Tripathi M, Kaistha SD. Hexavalent chromium reduction and plant growth promotion by Staphylococcus arlettae Strain Cr11. Chemosphere. 2012;86(8):847-852.
Crossref - Hu X, Chen J, Guo J. Two phosphate- and potassium-solubilizing bacteria isolated from Tianmu Mountain, Zhejiang, China. World J Microbiol Biotechnol. 2006;22(9):983-990.
Crossref - Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996;42(3):207-220.
Crossref - Karthik C, Arulselvi PI. Biotoxic effect of chromium (VI) on plant growth-promoting traits of novel Cellulosimicrobium funkei strain AR8 isolated from Phaseolus vulgaris rhizosphere. Geomicrobiol J. 2017;34(5):434-442.
Crossref - Sagar SS, Kumar R, Kaistha SD. Efficacy of phage and Ciprofloxacin Co-therapy on the formation and eradication of Pseudomonas aeruginosa biofilms. Arab J Sci Eng. 2017;42(1):95-103.
Crossref - Zablocka-Godlewska E, Przystas W, Grabinska-Sota E. Dye decolourisation using two Klebsiella strains. Water Air Soil Pollut. 2015;226:2249.
Crossref - Aksu Z, Kilic NK, Ertugrul S, Donmez G. Inhibitory effects of chromium(VI) and Remazol Black B on chromium(VI) and dyestuff removals by Trametes versicolor. Enzyme Microb Technol. 2007;40(5):1167-1174.
Crossref - Wuhrmann K, Mechsner K, Kappeler T. Investigation on rate – Determining factors in the microbial reduction of azo dyes. Eur J Appl Microbiol Biotechnol. 1980;9(4):325-338.
Crossref - Leelakriangsak M, Borisut S. Characterization of the decolorizing activity of azo dyes by Bacillus subtilis azoreductase AzoR1. Songklanakarin J Sci Technol. 2012;34(5):509-516.
- Chang JS, Chou C, Lin YC, Lin PJ, Ho JY, Hu TL. Kinetic characteristics of bacterial azo-dye decolorization by Pseudomonas luteola. Water Res. 2001;35(12):2841-2850.
Crossref - Garg SK, Tripathi M, Singh SK, Tiwari JK. Biodecolorization of textile dye effluent by Pseudomonas putida SKG-1 (MTCC 10510) under the conditions optimized for monoazo dye orange II color removal in simulated minimal salt medium. Int Biodeterior Biodegrad. 2012;74:24-35.
Crossref - Ogugbue CJ, Sawidis T, Oranusi NA. Evaluation of colour removal in synthetic saline wastewater containing azo dyes using an immobilized halotolerant cell system. Ecol Eng. 2011;37(12):2056-2060.
Crossref - Meng X, Liu G, Zhou J, Shiang Fu Q, Wang G. Azo dye decolorization by Shewanella aquimarina under saline conditions. Bioresour Technol. 2012;114:95-101.
Crossref - Kadam AA, Kulkarni AN, Lade HS, Govindwar SP. Exploiting the potential of plant growth promoting bacteria in decolorization of dye Disperse Red 73 adsorbed on milled sugarcane bagasse under solid state fermentation. Int Biodeterior Biodegrad. 2014;86:364-371.
Crossref - Parshetti GK, Telke AA, Kalyani DC, Govindwar SP. Decolorization and detoxification of sulfonated azo dye methyl orange by Kocuria rosea MTCC 1532. J Hazard Mater. 2010;176:503-509.
Crossref - Kumari L, Tiwary D, Mishra PK. Biodegradation of C.I. Acid Red 1 by indigenous bacteria Stenotrophomonas sp. BHUSSp X2 isolated from dye contaminated soil. Environ Sci Pollut Res. 2016;23(5):4054-4062.
Crossref - Chaudhari AU, Tapase SR, Markad VL, Kodam KM. Simultaneous decolorization of reactive Orange M2R dye and reduction of chromate by Lysinibacillus sp. KMK-A. J Hazard Mater. 2013;262:580-588.
Crossref - Ahemad M. Enhancing phytoremediation of chromium-stressed soils through plant-growth-promoting bacteria. J Genet Eng Biotechnol. 2015;13(1):51-58.
Crossref - Bhardwaj G, Shah R, Joshi B, Patel P. Klebsiella pneumoniae VRE36 as a PGPR isolated from Saccharum officinarum cultivar Co99004. J Appl Biol Biotechnol. 2017;5(01):47-52.
Crossref - Das N, Basak LVG, Salam JA, Abigail EA. Application of biofilms on remediation of pollutants – an overview. J Microbiol Biotechnol Res. 2017;2(5):783-790.
- Prakash B, Veeregowda BM, Krishnappa G. Biofilms: A survival strategy of bacteria. Curr Sci. 2003;85(9):1299-1307.
- Shafqat M, Khalid A, Mahmood T, Siddique MT, Han JI, Habteselassie MY. Evaluation of bacteria isolated from textile wastewater and rhizosphere to simultaneously degrade azo dyes and promote plant growth. J Chem Technol Biotechnol. 2017;92(10):2760-2768.
Crossref - Shahid M, Mahmood F, Hussain S, et al. Enzymatic detoxification of azo dyes by a multifarious Bacillus sp. strain MR-1/2-bearing plant growth-promoting characteristics. 3 Biotech. 2018;8:425.
Crossref - Al-Tohamy R, Sun J, Fareed MF, Kenawy ER, Ali SS. Ecofriendly biodegradation of Reactive Black 5 by newly isolated Sterigmatomyces halophilus SSA1575, valued for textile azo dye wastewater processing and detoxification. Sci Rep. 2020;10:12370.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.