ISSN: 0973-7510
E-ISSN: 2581-690X
Microbial utilization of uncommon C4 dicarboxylate L-tartrate is largely anaerobic, with aerobic L-tartrate utilization known for few bacterial species including Rhodopseudomonas sphaeroides and Pseudomonas putida. Aerobic L-tartrate-utilizing microbes could be industrially relevant owing to the efficient nature of the bioprocess and catalytic versatility of tartrate dehydrogenase (TDH) responsible for aerobic catabolism of L-tartrate. Present work involves isolation and characterization of Bacillus strains capable of aerobic L-tartrate utilization and its correlation with occurrence of TDH activity. Two out of 37 isolates, IC1-G and IC1-Y were identified as Bacillus megaterium spp. showing efficient aerobic growth, utilizing ~3.7 and 2.8 mM L-tartrate respectively at the end of 48 h. Several organic acids possibly including oxalic, succinic and citric acids were secreted as by-products of L-tartrate metabolism. Utilization of L-tartrate directly correlated with induction of TDH activity by ~3.2 and 5.2 folds in IC1-G and IC1-Y respectively, when grown in presence of L-tartrate as compared to when grown on citrate. Overall, this study contributes Bacillus as only the third genus capable of aerobic, TDH mediated L-tartrate utilization. These Bacillus isolates thus offer potential targets to develop an industrially relevant bioprocess and biocatalyst.
L-Tartrate; Bacillus species; Tartrate dehydrogenase.
Microorganisms display extensive metabolic adaptability to sustain the changes in their environment, especially with respect to availability of wide range of carbon sources and oxygen. Such evolution of metabolic capacity could be largely achieved through genetic mutations or horizontal gene transfers that add new enzyme-encoding genes as well as by modulating the metabolic fluxes, gene expression and regulation of existing enzymes1. When much preferred carbon sources like sugars and related derivatives are unavailable, bacteria utilize C4 dicarboxylates as carbon sources commonly including succinate, L-malate, oxaloacetate or aspartate, either aerobically through citric acid cycle or anaerobically via fumarate respiration (fumarate used as an electron acceptor and reduced to succinate) 2. Therefore, transporters, enzymes and regulations involved in C4-dicarboxylate metabolism under aerobic and anaerobic conditions are entirely different and have been extensively characterized in E. coli 3.
Microbial utilization of uncommon C4 dicarboxylates like tartrate via both aerobic and anaerobic routes has been experimentally supported, although is known to be mediated by distinct enzymes. Microbes including Aerobacter aerogenes, Clostridium sp., E. coli, Ruminococcus pasteurii and Ilyobacter tartaricus4-6 catabolize tartrate anaerobically via tartrate dehydratase (EC 4.2.4.32), catalyzing the conversion of tartrate to oxaloacetate which gets further metabolized by fumarate respiration. In related bacterial species like Klebsiella, anaerobic tartrate utilization however occurs via a different route involving Na+-dependent oxaloacetate decarboxylase3. Distinct dehydratases exist for each optical isomer, L-, D- and meso-tartrate, which are induced only by their specific substrates4. On the other hand, only few bacterial species, Rhodopseudomonas sphaeroides Y7, Pseudomonas putida and Pseudomonas acidovorans are known to aerobically metabolize L-tartrate, through tartrate dehydrogenase (TDH; EC 1.1.93) yielding oxaloglycolate8; with TDH possessing additional ability to catalyze oxidative decarboxylation of D-malate to pyruvate (R. sphaeroides) and meso-tartrate to D-glycerate (Pseudomonas sp.). Of the stereoisomers known, L-tartaric acid is most abundant in nature, found as chief component of grapes, bananas and tamarind. L-Tartrate also accumulates as a major by-product in wine industry and like other components in the winery waste it could be useful as potential feedstock for bioconversions to produce variety of industrially relevant platform compounds9. Therefore, microbial utilization of tartaric acid stereoisomers, specifically L-tartrate could be explored to develop industrially and pharmaceutically relevant bioprocesses like conversion of L-tartrate in winery waste to lactic and succinic acids6,9 as well as biocatalysts like tartrate decarboxylase or TDH (in conjunction with decarboxylating oxaloglycolate reductase) capable of converting L-tartrate into chiral synthon D-glycerate10.
In this regards, screening of aerobic L-tartrate utilizing microbes may prove more viable approach for two reasons (i) broad substrate specificity of TDH facilitating aerobic tartrate utilization, may help yielding diverse compounds like D-glycerate, dihydroxyfumarate or hydroxypyruvate from L- or meso-tartrate and (ii) aerobic conditions of growth facilitate rapid production of higher biomass, which if subsequently combined with production of target metabolites in anaerobic phase could help developing a more efficient bioprocess enabling maximum product yields at high cell densities. Typically, Bacillus spp. has been preferably explored in the present study owing its wide-spread acceptance as safe and robust bacteria suitable for development of new and improved industrially relevant processes. Thus, the present work attempts to isolate and characterize Bacillus spp. capable of aerobic L-Tartrate utilization and correlating it with occurrence of TDH activity.
Isolation, characterization and identification of tartrate-utilizing bacteria
Initially, soil sample was collected from randomly selected locations including domestic gardens and agricultural lands and stored at 4°C. These samples were collected across a period of 20 days and bacterial isolation was carried out within 3 days of sampling. For bacterial isolation, 1 g of soil sample was suspended in 10 ml of sterile normal saline aseptically followed by vigorous shaking at 100 rpm for 20 minutes. After allowing the soil particles to settle, the supernatant was serially diluted to prepare 10-2, 10-3, 10-4, 10-5 and 10-6 dilutions. For primary screening, 0.1 ml of each dilution was spread on nutrient agar plate and the colonies obtained after 24 h of incubation at 30°C and 37°C were used to screen for Bacillus species based on colony and cellular features, Gram’s staining and endospore staining. Selected colonies were further characterized and differentiated by growing on Hichrome Bacillus agar as per manufacturer’s instructions.
Tartrate utilization by bacterial isolates: Physiological experiments and analytical procedures
Single colony of selected isolates inoculated in 3 ml nutrient broth was allowed to grow with overnight shaking (150 rpm, 30 °C / 37 °C) and the cultures were directly used as bacterial inoculums. Freshly grown cultures (0.1 ml) were used to inoculate 20 ml of M9 minimal medium in a 150 ml conical flask, supplemented with 10 mM L-tartaric acid, 0.2 mM MgSO4. 6H2O, 0.1 mM CaCl2.6H2O and micronutrient cocktail (3.5 g/L FeSO4.7H2O, 0.16 g/L ZnSO4.7H2O, 0.08 g/L CuSO4.5H2O, 0.5 g/L H3BO3, 0.03 g/L CaCl2.2H2O,0.4 g/L MnSO4.4H2O) 11. With the initial cell density of ~0.02 (measured as OD600nm), the cultures were allowed to grow aerobically by shaking the flasks on an orbital rotary shaker (Orbitek, Scigenics Biotech Pvt. Ltd., India) maintained at 30 °C / 37 °C and 150 rpm up to 48 h. One milliliter of culture samples was aseptically withdrawn at regular time intervals to measure OD600 and were subsequently stored at -20°C until further analysis. Change in cell density (OD600) (UV-1800, Shimadzu Corp., Japan) was used as the measure of growth.
Stored samples were centrifuged at 8000xg for 5 min to derive cell free culture supernatant for measuring residual tartrate by ammonium metavanadate-based method12, where the absorbance of metavanadate-tartrate complex was recorded at 485 nm. Tartrate consumed was determined by deducting the value of residual tartrate measured in the spent medium from the tartrate concentration supplemented initially. Remaining supernatant was filtered through 0.2 µm nylon membranes for use in HPLC analysis (Waters Corp., USA). HPLC was performed by modified method using C-18 column operated at ambient temperature with 0.01 M K2HPO4 (pH 2.2, set using orthophosphoric acid) as mobile phase at a flow rate of 1.0 ml/min and the column effluents were monitored using a UV detector at 215 nm11. A 10 mM each of L-tartaric, oxalic, citric, malic, succinic acids and 5 mM of D-glycerate were used as organic acid standards since they are some of the commonly known intermediates and end-products of bacterial metabolism. Organic acids identified in the bacterial culture supernatants were quantified using area under the peak obtained for corresponding standard and were normalized with respect to cell density.
Molecular characterization of selected bacterial isolates
Identification of the selected tartrate utilizing isolates was carried out by 16S rDNA based molecular characterization. Genomic DNA from each bacterial isolate was isolated using standard procedure with desired modifications 13 and was used as template DNA in the polymerase chain reaction (PCR) for amplifying ~1500 bp region of 16S rDNA gene using universal primers 8F and 1492R. PCR reaction system contained ~100 ng of template DNA, 20 picomoles of each primer, 0.2 mM dNTPs, 1X PCR buffer and 1 U of Taq DNA polymerase (acquired from Takara Bio USA, Inc) in a total volume of 20 µl. Thermal cycler system (MJ Mini, Bio-Rad) was programmed as follows: initial denaturation at 94 °C for 5 min followed by 40 repeated cycles of primer annealing at 56 °C for 1 min, extension at 72 °C for 1 min and denaturation at 94 °C for 1 min, with a final extension step at 72 °C for 5 min. Resultant PCR amplicons were size-confirmed using 1 % agarose gel electrophoresis and PCR products of appropriate size were subjected to partial DNA sequencing using universal 8F primer, as an outsourced service (1st BASE, Singapore). The DNA sequences obtained were subsequently subjected to BLAST analysis for identification of the isolates (http://www.ncbi.nlm.nih.gov) based on DNA sequence similarities.
Preparation of cell free extracts and TDH enzyme assay
Selected isolates were cultivated on M9 minimal medium containing 10 mM L-tartrate as sole carbon source as described earlier and the cultures were harvested in mid-log to late-log growth phase by centrifugation at 8000xg for 5 minutes at 4 °C. The cell pellet was re-suspended in 1 ml of 0.1 M Tris-Cl (pH-8.5) buffer containing 1 % glycerol and treated with 100 µg / ml lysozyme for 10 min at room temperature. The cells were then subjected to sonication (VibraCellTM, SONICS, USA) for a total period of 3-5 minutes at a pulse rate of 10 s in an ice bath, followed by centrifugation at 8000xg at 4 °C for 30 minutes to remove cell debris. The resulting supernatant was used as the cell-free extract to determine TDH activity by following initial rate of NADH formation at 340 nm 7. The final volume of 1 ml assay system contained 100 mM Tris-Cl buffer (pH-8.5), 1 mM b-mercaptoethanol, 0.4 mM MnCl2, 50 mM (NH4)2SO4, 1.8 mM NAD and appropriate amount of crude cell lysate as enzyme source. The reaction was started by adding 50 mM L-tartrate (pH-8.5). Assay was carried out at room temperature (30 ± 2 °C) against appropriate controls lacking substrate and crude enzyme in the reaction mix. Total protein content of the cell lysate was measured using Biorad protein reagent as per manufacturer’s instruction, with bovine serum albumin as standard. One unit of enzyme activity was defined as amount of enzyme required to convert 1 µmole of substrate per minute per ml of crude enzyme per mg of total protein. Cell extracts of cultures similarly grown on M9 minimal medium containing 10 mM citrate as sole carbon source were also used to determine TDH activity for comparative analysis.
Statistical analysis
Data for quantitative experiments has been represented as Mean ± S.D as indicated in the respective figure legends. Statistical significance between values compared was derived using Student’s t-test performed using Microsoft Excel.
Isolation, characterization and identification of tartrate-utilizing bacteria
Primary screening of bacterial colonies isolated from randomly collected soil samples was based on their microbial characteristics like Gram’s and endospore staining as well as typical cell shape, yielding about 37 Bacillus-like isolates. Distinct colonies from diverse source soil samples were considered while screening. Comparative analysis of detailed colony morphology and features as observed on nutrient agar as well as characteristic pigmentation profile on differential HiChrome Bacillus agar indicated that most isolates were found distinct while some of the closely similar isolates could be differentiated based on their pigmentation.
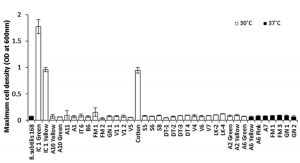
These 37 isolates were subjected to aerobic growth on defined minimal medium containing L-tartrate as sole carbon source. At the end of 48 h only 3 isolates viz IC1-G, IC1-Y and C-1, managed to show significant growth on tartrate, attaining maximum cell densities of ³ 1.0 OD600nm (Fig.1). Remaining isolates, including type strain B. subtilis 168 (IA1) used as negative control, failed to grow on tartrate as indicated by their maximum cell densities not exceeding 0.15 OD600nm, even after 48 h of incubation, suggesting poor tartrate utilizing ability. Molecular identification of these three tartrate-utilizing isolates based on partial 16S rDNA sequencing identified IC1-G and IC1-Y as Bacillus megaterium species while C-1 was identified as Bacillus endophyticus (Table 1). Repeated attempts to grow on tartrate revealed that C-1 isolate failed to show consistent growth under shaking conditions and hence was omitted from subsequent studies.
Fig. 1. Relative abilities of primarily screened bacterial isolates to grow on defined minimal medium containing L-tartrate as sole carbon source
Maximum cell densities were measured after 48 h of incubation under aerobic condition at respective temperature of isolation.
Results are depicted as Mean ± SD of 3-4 independent observations
Table (1):
Partial 16S rDNA sequencing-based molecular identification of tartrate utilizing bacterial isolates.
Sr. No |
Isolate |
Query length (bp) |
Query coverage (%) |
Homology (%) |
Identified species |
GenBank Accession number |
|---|---|---|---|---|---|---|
1 |
IC1-Y |
973 |
99 |
99.38 |
Bacillus megaterium |
MN080445 |
2 |
IC1-G |
937 |
95 |
97.54 |
Bacillus megaterium |
MN080444 |
3 |
C-1 |
1362 |
99 |
97.93 |
Bacillus endophyticus |
MN080446 |
Tartrate utilization by IC1-G and IC1-Y isolates
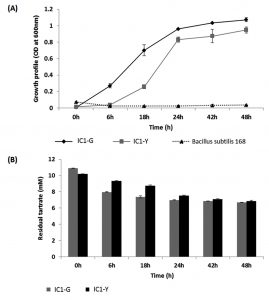
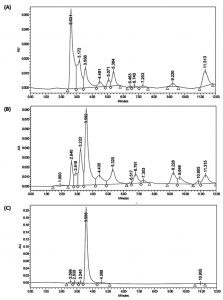
Systematic growth curve of IC1-G and IC1-Y isolates performed by growing them on M9 minimal medium with tartrate as sole carbon source under shaking conditions revealed that both the isolates began attaining stationary phase after 24 h of growth (Fig. 2A). IC1-G showed relatively better growth on tartrate as compared to IC1-Y. This growth could be attributed to utilization of ~3.7 and 2.8 mM tartrate utilization by IC1-G and IC1-Y respectively at the end of 48 h (Fig. 2B). Apparently major tartrate utilization (more than 90 % of the total) was achieved at the end of 24 h only. HPLC profile of the supernatants of 48 h old tartrate-grown cultures of IC1-G and IC1-Y when compared to that of 0 h growth medium containing tartrate, revealed a range of organic acids secreted as by-products of aerobic tartrate metabolism. IC1-G secreted 0.42 ± 0.18 µg/ml/cell OD600nm of oxalic acid (Rt ~7.29) while IC1-Y secreted 0.95 ± 0.19 and 7.11 ± 3.16 µg/ml / cell OD600nm of oxalic (Rt ~7.3) and citric acids (Rt ~10.86), respectively (Fig. 3). Oxalic and citric acid standards used for quantitation were retained at 7.13 and 10.75 minutes respectively. A prominent peak detected at Rt ~11.3 could be that of succinic acid however it could not be confirmed. Other organic acids detected in the HPLC profiles remain unidentified.
Fig. 2. Aerobic growth and L-tartrate utilization profile of selected isolates as compared to standard strain Bacillus subtilis 168
(A) Growth profile on M9 minimal medium containing L-tartrate as sole carbon source and (B) Residual tartrate levels as
monitored in 48 h old culture supernatants. Results are depicted as mean ± SD of 3-4 independent observations
Fig. 3. HPLC analysis of organic acids secreted by selected isolates as by-products of aerobic L-tartrate metabolism Representative HPLC chromatograms of cell free culture supernatants of (A) IC1-G and (B) IC1-Y isolates grown for 48 h on L-tartrate as sole carbon source (C) initial 0 h growth medium containing tartrate
Correlation of tartrate utilization by Bacillus isolates with occurrence of TDH
Aerobic assimilation of tartrate usually involves conversion of tartrate to common metabolic intermediate glycerate, using TDH and oxaloglycolate reductase or tartrate dehydrogenase/decarboxylase as known in Pseudomonas species 3. Accordingly, occurrence of functional TDH in tartrate-utilizing Bacillus isolates IC1-Y and IC1-G was monitored independently in cells grown on citrate and L-tartrate as sole carbon sources. Citrate was used as the control substrate since it is a common organic acid aerobically transported and metabolized via mechanisms similar to those for L-tartrate and thus could serve as appropriate reference to indicate specific tartrate-mediated induction of TDH. In presence of L-tartrate as sole carbon source, specific activities of TDH in IC1-G and IC1-Y were respectively induced by ~3.2 and 5.2 folds, as compared to that in citrate-grown cells (Table 2). These results suggested that tartrate metabolism by IC1-G and IC1-Y clearly correlated with TDH activity, which was specifically induced by the presence of tartrate. On the other hand, while basal level of TDH activity detected in citrate-grown cells of IA1 was comparable to similarly grown cells of IC1-G and IC1-Y, induction of TDH in type strain IA1 could not be monitored under experimental conditions owing to lack of its growth on tartrate as sole carbon source.
Table (2):
Occurrence of inducible tartrate dehydrogenase activity in L-tartrate-utilizing Bacillus isolates.
| No | Strains | Carbon source used for cell growth | Specific TDH activity (µmoles/min/ml/mg total protein) |
|---|---|---|---|
| 1 | IA 1
Bacillus subtilis 168 |
Tartrate | Not detected |
| Citrate | 10.23 ± 3.58 | ||
| 2 | IC1-G
(Bacillus megaterium) |
Tartrate | 58.49 ± 6.18 *** |
| Citrate | 18.33 ± 3.42 | ||
| 3 | IC1-Y
(Bacillus megaterium) |
Tartrate | 63.39 ± 1.09 *** |
| Citrate | 12.01 ± 1.71 |
Results are expressed as Mean ± SD of 3-5 independent observations; ***p<0.001as compared to citrate-grown control cells. IA1 failed to grow on L-tartrate as sole carbon source
Metabolism of C4-dicarboxylate under both aerobic and anaerobic growth conditions has been extensively studied in E. coli 14, with major focus on mechanisms of their transport and regulation of the metabolic pathways at the transcriptional level. With respect to uncommon C4-dicarboxylates like L-tartrate, anaerobic uptake and metabolism has been investigated while the detailed mechanisms of its aerobic utilization in E. coli are largely unclear. In fact, microbes capable of aerobic utilization of L-tartrate are relatively underexplored despite having an obvious advantage with respect to development of industrial bioprocesses. Considering the established relevance of Bacillus genus in producing industrially important bioproducts and bioprocesses, the present work aimed at isolating Bacillus species capable of aerobic L-tartrate utilization and investigating the occurrence of associated characteristic enzyme TDH.
Initial microbial characterization and subsequent molecular characterization identified two isolates as Bacillus megaterium species, capable of consistent utilization of L-tartare under fully aerated conditions. Our results with these two isolates IC1-G and IC1-Y suggested that they could utilize around 37 and 29% of total L-tartrate supplemented as carbon source. Bacillus species have been earlier reported to show anaerobic L-tartrate degradation in addition to several other bacterial species6,15 ; however, this is probably the first study reporting aerobic L-tartrate utilization by Bacillus sp. As such, anaerobic tartrate fermenting bacteria outnumber the aerobic tartrate utilizers, of which L-tartrate fermenters are much more prevalent than the D- and meso-tartrate fermenters. In this scenario, the present study not only adds to the pool of aerobic tartrate utilizers but could also prove useful in facilitating the exploration of these microbes for enhancing the fundamental understanding of tartrate metabolism as well as its industrial applications.
Efficient growth of IC1-Y and IC1-G isolates on L-tartrate suggested presence of both functional tartrate uptake as well as catabolic mechanisms; absence of either of which could be responsible for lack of tartrate utilization by IA1 and other isolates of Bacillus species even after 48 h of incubation. Bacterial metabolism of L-tartrate is known to require induction of specific transporters and catabolic enzymes. Anaerobic uptake of L-tartrate is mediated by ttdT encoded tartrate-succinate antiporter followed by catabolism via L-tartrate dehydratase; their genes being jointly organized in an operon (TtdAB) 16. On the other hand, aerobic L-tartrate transport is usually mediated by dctA encoded succinate carrier functional at neutral pH3,14. While the present study does not clearly explain the mechanisms involved in L-tartrate uptake by IC1-Y and IC1-G, a similar C4-dicarboxylate transporter DctA has been annotated in Bacillus megaterium genome (BG04_RS04505; Gene ID: 29910369), which could be anticipated to play a role in L-tartrate transport. Although uptake of tartrate has been experimentally studied in Bacillus subtilis, its mechanisms and kinetics are quite unclear17. Following the uptake, intracellular aerobic breakdown of L-tartrate could then occur via TDH and oxaloglycolate reductase, or tartrate decarboxylase, yielding glycerate as the product as reported for several Pseudomonas sp.18-20. Alternatively, a bifunctional L-tartrate dehydrogenase-D-malate dehydrogenase, also capable of catalyzing conversion of meso-tartrate to glycerate, has been reported in Rhodopseudomonas sphaeroides while D-malate dehydrogenase of isocitrate and isopropylmalate dehydrogenases family is known to be induced by the presence of tartrate stereoisomers21. In this regard, even TDH is considered to be the evolutionary descendent of isopropylmalate dehydrogenase, probably acquired by bacteria to manage L-tartrate catabolism under environmental pressures8,22. Notably, ycsA gene and BG04_RS21075 (Gene ID: 29907790) are already annotated as TDH encoding genes in B. subtilis and B. megaterium genomes respectively. Accordingly, significant levels of TDH activity detected in crude lysates of L-tartrate grown cultures of IC1-Y and IC1-G, suggested TDH to be the prime mechanism for L-tartrate catabolism in Bacillus megaterium species. Correspondingly, basal level TDH activity in citrate-grown cultures of IC1-Y and IC1-G indicated that TDH activity was specifically induced by L-tartrate; induction of which by other stereoisomers of tartrate however has not been explored in the present study.
Secretion of a range of metabolic by-products mostly organic acids, by IC1-G and IC1-Y was demonstrated using HPLC profiling of culture supernatants confirming successful utilization of tartrate and its metabolism. However, based on the array of standard organic acids used in the present study, most organic acids remained unidentified, except oxalic and citric acids; the metabolic basis for the formation of which remain unclear. Acetate, formate, succinate, lactate and carbon dioxide are some of the known products of anaerobic L-tartrate fermentation6. On the other hand, while aerobic tartrate utilization in few Pseudomonas species including P. putida is well characterized, their metabolic by-products are not clearly known.
Overall our results report Bacillus as only third genus after Pseudomonas and Rhodopseudomonas, capable of aerobic tartrate utilization, in a pathway mediated by TDH. Additionally, owing to the well-known broad substrate specificity of TDH as known from P. putida8, detailed biochemical characterization of TDH from Bacillus spp. could further offer a promising target to develop biocatalyst capable of producing industrially relevant biomolecules.
ACKNOWLEDGEMENTS
Not applicable.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DP has contributed in execution of the experiments, data recording, compilation and drafting manuscript. AB has contributed in planning of experiments, data analysis, conceptualizing, writing and proof-reading of the manuscript.
FUNDING
The work has been funded by Science and Engineering Research Board, Department of Science & Technology, Government of India [Grant numbers: SR/FT/LS-92/2012; EMR/2016/003524].
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are available from the corresponding author, [A. Buch], upon request. The partial 16S rDNA sequences are submitted to GenBank and the accession numbers are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Rodrigues JF, Wagner A. Evolutionary plasticity and innovations in complex metabolic reaction networks. PLoS Comput Biol., 2009; 5(12): e1000613.
Crossref - Wosten MM, van de Lest CH, van Dijk L. Function and regulation of the C4-dicarboxylate transporters in Campylobacter jejuni. Front Microbiol., 2017; 8: 174.
Crossref - Unden G, Strecker A, Kleefeld A, Kim OB. C4 -dicarboxylate utilization in aerobic and anaerobic growth. EcoSal Plus, 2016; 7(1).
Crossref - Shilo M, Stanier RY. The utilization of the tartaric acids by Pseudomonads. Microbiol., 1957; 16(2): 482–490.
Crossref - Rode H, Giffhorn F. D-(-)-Tartrate dehydratase of Rhodopseudomonas sphaeroides: purification, characterization, and application to enzymatic determination of d-(-)-tartrate. J. Bacteriol., 1982; 150(3): 1061–1068.
- Schink B. Fermentation of tartrate enantiomers by anaerobic bacteria, and description of two new species of strict anaerobes, Ruminococcus pasteurii and Ilyobacter tartaricus. Arch Microbiol., 1984; 139(4): 409–414.
Crossref - Giffhorn FR, Kuhn AN. Purification and characterization of a bifunctional L- (+) -tartrate dehydrogenase-D-(+)-malate dehydrogenase (dcarboxylating) from Rhodopseudomonas sphaeroides. Y. J. Biotechnol., 1983; 155(1): 281–290.
- Malik R, Viola RE. Structural characterization of tartrate dehydrogenase: A versatile enzyme catalyzing multiple reactions. Acta Crystallogr. D. Struct. Biol., 2010; 66(6): 673–684.
Crossref - Zacharof MP. Grape winery waste as feedstock for bioconversions: applying the biorefinery concept. Waste Biomass Valorization, 2017; 8(4): 1011–1025.
Crossref - Furuyoshi S, Kawabata N, Tanaka H, Soda K. Enzymatic production of D-glycerate from L-tartrate. Agric. Biol. Chem., 1989; 53(8): 2101-2105.
Crossref - Buch A, Archana G, Naresh Kumar G. Enhanced citric acid biosynthesis in Pseudomonas fluorescens ATCC 13525 by overexpression of the Escherichia coli citrate synthase gene. Res. Microbiol., 2008; 159(9-10): 2620–2629.
Crossref - Zoecklein B, Fugelsang KC, Gump B, Nury FS. Wine analysis and production. Springer US. https://doi.org/10.1007/978-1-4757-6967-8
- Shahriar M, Haque MR, Kabir S, Dewan I, Bhuyian MA. Effect of proteinase-K on genomic DNA extraction from gram-positive strains. Stamford J. Pharm. Sci., 2011; 4(1): 53–57.
Crossref - Lukas H, Reimann J, Kim OB, Grimpo J, Unden G, Regulation of aerobic and anaerobic D-malate metabolism of Escherichia coli by the LysR-type regulator DmlR (YeaT). J. Bacteriol., 2010; 192(10): 2503–2511.
Crossref - Kampitz LO, Lynen F. Formation of oxaloacetate from D-tartrate. Proc World Cong Neurol Surg World Fed. Neurosurg. Soc., 1956; 5: 292-293.
- Kim OB, Reimann J, Lukas H, Schumacher U, Grimpo J, Unden G. Regulation of tartrate metabolism by TtdR and relation to the DcuS – DcuR-regulated C 4 – dicarboxylate metabolism of Escherichia coli. Microbiol., 2009; 155(11): 3632–3640.
Crossref - Fournier RE, Pardee AB. Evidence for inducible, L-malate binding proteins in the membrane of Bacillus subtilis. Identification of presumptive components of the C4-dicarboxylate transport system. J. Biochem., 1947; 249(18): 5948-5954.
- Kohn LD, Jakoby WB. Tartaric Acid Metabolism. VI. Crystalline oxaloglycolate reductive decarboxylase. J. Biol. Chem., 1968; 243(10): 2486–2493.
- Furuyoshi S, Nawa Y, Kawabata N, Tanaka H. Purification and characterization of a new NAD+ -dependent enzyme, L-tartrate decarboxylase, from Pseudomonas sp. group Ve-2. J. Biochem., 1991; 110(4): 520–525.
Crossref - Reaney SK, Begg C, Bungard SJ, Guest JR. Identification of the L-tartrate dehydratase genes (ttdA and ttdB) of Escherichia coli and evolutionary relationship with the Class I fumarase genes. Microbiol., 1993; 139(7): 1523–1530.
Crossref - Ebbighausen H, Giffhorn F. A novel mechanism involved in the metabolism of the tartaric stereoisomers in Rhodopseudomonas sphaeroids : enzymatic conversion of meso-tartaric acid to D-glyceric acid and CO2. Arch. Microbiol., 1(4): 338-344.
Crossref - Tipton PA, Beecher BS. Tartrate dehydrogenase a new member of the family of metal-dependent decarboxylating R-hydroxyacid dehydrogenases. Arch. Biochem. Biophys., 1994; 313(1): 15-21.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.