ISSN: 0973-7510
E-ISSN: 2581-690X
The antibiotic resistance pattern was observed significantly in various geographical locations. Routine surveillance is therefore essential for constant monitoring of AMR rates in the clinically important pathogens. It is imperative to track the changing resistance pattern over time, to guide proper therapeutic strategies to combat infections due to drug-resistant pathogens. This study aims to highlights the distribution of aerobic bacterial isolated from pus samples, and their susceptibility to different antibiotics collected during 2017 (July to December) in a tertiary care hospital. Nearly 637 clinical pus samples were received during July to December 2017 to the Department of Microbiology, Tertiary care hospital, Puducherry. Bacterial identification was performed using standard conventional biochemical tests and antibiotic susceptibility was carried out according to CLSI guidelines 2017 on each one of the aerobic bacterial isolates from the pus samples. Among the isolates 76.5% were Gram-negative bacilli (GNB) as well as 23.5% were Gram-positive cocci (GPC). The most common bacteria isolated were Pseudomonas spp 24.88% (108 in 434), followed by Escherichia coli 21.66% (94 in 434), Staphylococcus aureus 19.82% (86 in 434) and Klebsiella pneumoniae 13.13% (57 in 434). Of the 86 (19.82%) Staphylococcus aureus isolates, 16 (18.40%) were MRSA. Pseudomonas aeruginosa was highly susceptible to the carbapenems and least susceptible to ciprofloxacin. Acinetobacter baumannii was the most resistant organism according to this study and showed the least susceptibility to ceftriaxone and maximum susceptibility to aminoglycosides. This study concluded that the Pseudomonas aeruginosa isolate was found to be a predominant in our clinical pus samples. Gram negative bacteria are more commonly associated with the pyogenic lesion that Gram positive. A high level of an antibiotic resistance was observed in most of our bacterial isolates.
Pus, Antibiotic susceptibility, antimicrobial resistance, bacteriological profile
Pus is one of the most readily recognizable signs of an infection. Also traditionally defined as laudable pus is the thick, white, odorless exudate formed by pyogenic bacterial infections1. Both aerobic and anaerobic bacteria have been found in hospital acquired infections, especially post-operative wound infections resulting in substantial morbidity, prolonged hospital stay makes a layman to create an economic stress factors2. Antibiotics are widely used for therapeutic and prophylactic purposes but their unselective use in humans and animals combined with improved worldwide connectivity has led to a surge in antibiotic resistance3-5. Antimicrobial resistance (AMR) especially Gram-negative bacilli have emerged as a significant public health problem globally due to insufficient treatment options. Infections from resistant organisms are linked to increased mortality and economic costs5. India faces one of the world’s greatest burdens of drug-resistant pathogens6. The rate of resistance in Gram-negative organisms is higher than that of Gram-positives. High ESBL rates have been reported in E. coli, K. pneumoniae; increased colistin and carbapenems resistance in K. pneumoniae and high carbapenem resistance rates in Acinetobacter baumannii than in Pseudomonas aeruginosa7 Among Staphylococcus species a high inducible clindamycin resistance was mostly observed in MRSA compared to MSSA3. The resistance profile across different geographical locations varies significantly. Routine surveillance is therefore essential for constant monitoring of AMR rates in the clinically important pathogens3,8. Antibiotic sensitivity pattern has need to be monitored regularly for appropriate treatment required for multi-drug resistant pathogens. This study aims to highlights the distribution of aerobic bacterial isolated from pus samples, and their susceptibility to different antibiotics collected during 2017 (July to December) in a tertiary care hospital.
This Prospective study included 637 pus samples which were received in the Microbiology department during 2017 (July to December) at Mahatma Gandhi Medical College &Research Institute, Puducherry, India. Bacterial identification was performed using routine diagnostic tests viz., conventional biochemical (IMViC) and sugar fermentation tests to detect upto the level of species level4.The antibiogram of all the isolates was determined using Kirby-Bauer’s disk diffusion technique on Mueller-Hinton agar according to CLSI 2017 guidelines4. Antimicrobial agents tested were as follows: cephalosporin (cefoxitin, ceftazidime, and ceftriaxone); β-lactam/β-lactamase inhibitor (cefoperazone/sulbactam and piperacillin/tazobactam); carbapenems (imipenem and Meropenem); fluoroquinolones (ciprofloxacin); aminoglycosides (amikacin, gentamicin, tobramycin) and polymyxins (colistin and polymyxin B). Batch wise testing was made to check the Quality control (QC) for freshly prepared biochemicals as well as agar plates by using CLSI guidelines. For QC recommended bacterial strains were used viz., Escherichia coli ATCC 25922, and ATCC 25923 Staphylococcus aureus.
Statistical Analysis
The results obtained were analyzed using MS Excel, 2010 version, with counts, percentages and pivot tables.
Socio-demographic characteristics of the study population
Among 637 patients, 263 (41.29 %) were aged from 40 to 60 years followed by 203 (31.87 %) patients ranges from 20 to 39 years, 104 (16.33%) cases were aged more than 60 years and remaining 67 (10.51%) patients are below 20 years of age. In our study population, Male (56.36%) was the predominant when compared to females (43.80%) in our study population (Table 1) which might be explained by the fact that men are mostly involved in outdoor activities and occupation which increases their likelihood of injury for 71 (11.15%) samples were received from the Out-Patient Department (OPD) and (88.85%) from In-Patient Department (IPD).
Table (1):
Age & sex structure of the study population.
Age group |
Female |
Male |
Total (n=637) |
|---|---|---|---|
<20 |
26 (9.32%) |
41 (11.42%) |
67 (10.52%) |
20-39 |
92 (32.97%) |
111(30.92%) |
203 (31.86%) |
40-60 |
131(46.95%) |
132(36.77%) |
263 (41.29%) |
>60 |
30 (10.75%) |
74(20.61%) |
104 (16.333%) |
Total |
279 (100%) |
358 (99.8%) |
Bacterial Culture results for pus sample analysis
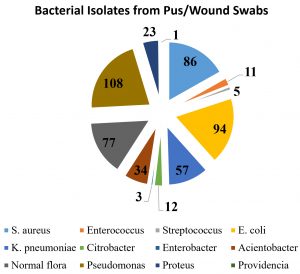
Of the 637 samples, 434 (72.81%) had bacterial profile from In-patient (IP – Ward) and out-patient (OPD) department, among these isolates, 332 (76.5%) were showed pathogenic various gram-negative bacilli and cocco-bacilli growth, 102 (23.5%) had Gram positive cocci viz., S. aureus, Enterococcus and Streptococcus sps. Remaining 203 (36.10%) cultures were sterile. Table 2 describes about the distribution of bacterial isolates from pus and wound samples. Nearly, 108 (24.88%) Pseudomonas isolates were found most predominant in pus samples followed by Escherichia coli (21.66%), Staphylococcus aureus (19.82%) which includes MRSA (18.40%) and Klebsiella pneumoniae (13.13%). Occurrence of other isolates in descending order are Acinetobacter sp (7.83%), Proteus mirabilis (5.30%), Citrobacter spp (2.76%), Enterobacter spp. (0.69%), Providencia (0.23%).
Table (2):
Distribution of Bacterial isolates from OP & IP patients (n=434).
S. No |
Name of the Isolate |
In-patient isolates (n=381) |
Out-patient isolates (n=53) |
Total (n=434) % |
|---|---|---|---|---|
1 |
Staphylococcus aureus |
73 (19.2%) |
13 (24.5%) |
86 (19.8%) |
2 |
Enterococcus sps. |
9 (2.4%) |
2 (3.8%) |
11 (2.5%) |
3 |
Streptococcus sps. |
5 (1.3%) |
0 |
5 (1.3%) |
4 |
E. coli |
86 (22.5%) |
8 (15.1%) |
94 (21.6%) |
5 |
K. pneumoniae |
52 (13.6%) |
5 (9.4%) |
57 (13.1%) |
6 |
Enterobacter sps. |
3 (0.8%) |
0 |
3 (0.7%) |
7 |
Citrobacter sps. |
9 (2.4%) |
3 (5.6%) |
12 (2.8%) |
8 |
Acinetobacter sps. |
31 (8.3%) |
3 (5.6%) |
34 (7.8%) |
9 |
Pseudomonas aeruginosa |
91 (23.9%) |
17 (32.1%) |
108 (24.9%) |
10 |
Proteus sps. |
21 (5.5%) |
2 (3.8%) |
23 (5.3%) |
11 |
Providencia sps. |
1 (0.3%) |
0 |
1 (0.3%) |
In this study, 11 bacterial pathogens were isolated from pus and wound swabs which includes S. aureus, E. coli, P. aeruginosa, K. pneumoniae, Citrobacter spp. Enterobacter spp., Enterococcus spp., Streptococcus spp. Proteus spp and Providencia spp. The majority of the specimen yielded gram-negative bacterial isolates (76.5%) which is superior to gram-positive bacteria (16.0%) and this has been seen in earlier studies8-10. The most common isolate was Pseudomonas aeruginosa, which coincided with findings of Lockhart et al and Agnihotri et al.11,12. Various researchers found that the S. aureus yields a predominant growth nearly 40–60% were isolated from different wound infection13-16. This difference in the distribution of bacterial isolates may be attributed to the differences in study design, type of lesion, geographical location and climatic conditions. Among the 86 (19.82%) S. aureus isolates, 16 (18.40%) were identified as MRSA due to the resistance of Cefoxitin drugs which acts as a surrogative marker. Almost similar findings were found in another study from Nepal that the most of the bacterial isolates in pus samples14. An efficient infection control program was followed and this could be results of reduced rate MRSA infections. Highest rate of susceptibility (95-100%) was seen toward teicoplanin, vancomycin and linezolid among the Gram-positive cocci. Both S. aureus and Enterococcus spp has been demonstrated that least sensitivity was observed towards ciprofloxacin (26-27%) (Table 3). In the earlier studies, gram-positive bacteria were predominant in most of the wound infections, but in contrast to the present study, gram negative bacteria i.e. P. aeruginosa was found frequently14. Hanumanthappa et al, from Ballari district that demonstrate yielded nearly 56% of positive culture, but it was low (68.2%) when compared to the present study9. According to the results of antibiogram of the present study, Pseudomonas aeruginosa was highly susceptible to the carbapenems (Meropenem followed by Imipenem) and least sensitive to ciprofloxacin (Table 3). A. baumannii was the most multi-drug resistant organism according to this study and showed a least sensitive to ceftriaxone and maximum to aminoglycosides (gentamicin followed by amikacin). E. coli isolates yields maximum sensitive to amikacin followed by meropenem, while being least susceptible to ampicillin and ceftriaxone. Proteus mirabilis did not exhibit any resistance to meropenem, piperacillin-tazobactum and cefaperazone-sulbactum but showed maximum resistance to ciprofloxacin and cotrimoxazole. The Providencia spp. isolated in this study was a pan sensitive strain. K. pneumoniae was more susceptible to tested antibiotics compared to Citrobacter spp. Both species showed resistance to the group of cephalosporins. Enterobacter isolates were highly susceptible to most of the broad spectrum of antibiotics.
Table (3):
Antibiotic susceptibility profile of the aerobic bacterial isolates from pus.
Antibiotic |
Staphylococcus aureus |
Enterococcus spp |
β hemolytic Streptococcus spp |
Escherichia coli |
Klebsiella pneumoniae |
Citrobacter spp |
Enterobacter spp |
Proteus mirabilis |
Providencia spp |
Acinetobacter spp |
Pseudomonas spp |
|---|---|---|---|---|---|---|---|---|---|---|---|
Ampicillin |
– |
54.5 |
80 |
21.3 |
– |
– |
– |
– |
– |
– |
– |
Penicillin |
53.4 |
63.6 |
80 |
– |
– |
– |
– |
– |
– |
– |
– |
Amikacin |
– ` |
84.0 |
78.95 |
83.33 |
100 |
82.6 |
100 |
52.9 |
75 |
||
Gentamycin |
95.3 |
71.3 |
64.91 |
50.00 |
66.6 |
73.9 |
100 |
55.8 |
69.4 |
||
Tobramycin |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
78.7 |
High level Gentamicin |
– |
81.8 |
– |
– |
– |
– |
– |
– |
– |
– |
– |
Clindamycin |
88.3 |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
Imipenem |
– |
75.5 |
73.68 |
58.33 |
100 |
78.2 |
100 |
50.0 |
83.3 |
||
Meropenem |
– |
82.9 |
78.95 |
75.00 |
100 |
100 |
100 |
47.1 |
82.4 |
||
Piperacillin-Tazobactum |
– |
73.4 |
73.68 |
83.33 |
100 |
100 |
100 |
44.1 |
81.5 |
||
Ciprofloxacin |
26.7 |
27.2 |
– |
28.7 |
49.12 |
33.33 |
66.6 |
39.1 |
100 |
41.2 |
51.8 |
Cotrimoxazole |
68.6 |
– |
– |
40.4 |
47.37 |
66.67 |
100 |
39.1 |
100 |
47.1 |
– |
Cefoperazone-sulbactum |
– |
– |
– |
78.7 |
71.93 |
75.00 |
100 |
100 |
100 |
50.0 |
79.6 |
Ceftriaxone |
– |
– |
– |
21.3 |
42.11 |
58.33 |
33.3 |
56.5 |
– |
14.7 |
– |
Ceftazidime |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
71.3 |
Cefoxitin |
81.4 |
– |
– |
– |
– |
— |
– |
– |
– |
– |
– |
Erythromycin |
59.3 |
45.4 |
40 |
– |
– |
– |
– |
– |
– |
– |
– |
Tetracycline |
48.8 |
36.3 |
40 |
– |
– |
– |
– |
– |
– |
– |
– |
Teicoplanin |
95.3 |
100 |
100 |
– |
– |
– |
– |
– |
– |
– |
– |
Vancomycin |
100 |
100 |
100 |
– |
– |
– |
– |
– |
– |
– |
– |
Linezolid |
100 |
100 |
100 |
– |
– |
– |
– |
– |
– |
– |
– |
To conclude that the gram-negative bacilli is more predominant when compared to gram positive and mostly associated with pyogenic lesions. The bacteriological profile of wound infections was similar to children as well as adults. A level of resistance was increasing in most of the bacterial isolates, this may reflect due to poor antibiotic stewardship. For monitoring the changing trend, periodic review of the bacteriological profile and pattern of antibiotic sensitivity is extremely essential.
Limitations
Isolation of anaerobic bacteria could not be carried out in the study and due to resource constraints, we were unable to confirm our results using molecular analysis. A multicenter study including larger sample size would have increased the significance of this study.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Research conception & design: Dr. NKB, Dr. US, Performing the experiments: Ms. DA, Data acquisition: Ms. DA, Dr. NKB, Dr. US, Data analysis and interpretation: Dr. NKB, Ms. D, Dr. US, Drafting of the manuscript: Dr. NKB, Ms. D, Critical revision of the manuscript: Dr. NKB, Ms. D, Dr. US.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
Data available in the Department of Microbiology, Mahatma Gandhi Medical College & Research Institute, Pondicherry. Could be provided if necessary to anyone.
- Freiberg JA. The mythos of laudable pus along with an explanation for its origin. J Community Hosp Intern Med Perspect. 2017;13;7(3):196-198.
Crossref - Bowler PG, Duerden BI, Armstrong DG. Wound Microbiology and Associated Approaches to Wound Management. Clin Microbiol Rev. 2001;14(2):244-269
Crossref - Veeraraghavan B, Walia K. Antimicrobial susceptibility profile & resistance mechanisms of Global Antimicrobial Resistance Surveillance System (GLASS) priority pathogens from India. Indian J Med Res. 2019;149(2):87-96.
Crossref - Wayne, PA. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. 2017. (CLSI supplement M100.).
- Vasoo S, Barreto JN, Tosh PK. Emerging Issues in Gram-Negative Bacterial Resistance: an update for the practicing clinician. Mayo Clin Proc. 2015;90(3):395-403.
Crossref - Dixit A, Kumar N, Kumar S, Trigun V. Antimicrobial resistance: Progress in the decade since emergence of New Delhi metallo-β-lactamase in India. Indian J Community Med. 20191;44(1):4
- Bonomo RA, Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis. 2006;43(Suppl 2):S49-S56.
Crossref - Trojan R, Razdan L, Singh N. Antibiotic Susceptibility Patterns of Bacterial Isolates from Pus Samples in a Tertiary Care Hospital of Punjab, India. Int J Microbiol [Internet]. 2016.
Crossref - Hanumanthappa P, Vishalakshi B, Krishna S. A Study on aerobic Bacteriological profile and Drug sensitivity pattern of Pus samples in a tertiary care hospital. Int J Curr Microbiol Appl Sci. 2016;5(1):95-102.
Crossref - Ghosh A, Karmakar PS, Pal J, Chakraborty N, Debnath NB, Mukherjee JD. Bacterial incidence and antibiotic sensitivity pattern in moderate and severe infections in hospitalized patients. J Indian Med Assoc. 2009;107(1):21-2.
- Lockhart SR, Abramson MA, Beekmann SE, et al. Antimicrobial Resistance among Gram-Negative Bacilli Causing Infections in Intensive Care Unit Patients in the United States between 1993 and 2004. J Clin Microbiol. 2007;45(10):3352-3359.
Crossref - Agnihotri N, Gupta V, Joshi RM. Aerobic bacterial isolates from burn wound infections and their antibiograms–a five-year study. Burns J Int Soc Burn Inj. 2004;30(3):241-243.
Crossref - Sawdekar H, Sawdekar R, Wasnik VR. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to antibiotic agents at super specialty hospital, Amravati city, India. Int J Res Med Sci. 2017;3(2):433-439.
Crossref - Rai S, Yadav UN, Pant ND, et al. Bacteriological Profile and Antimicrobial Susceptibility Patterns of Bacteria Isolated from Pus/Wound Swab Samples from Children Attending a Tertiary Care Hospital in Kathmandu, Nepal. Int J Microbiol. 2017;2017:1-5.
Crossref - Khanam RA, Islam MR, Sharif A, Parveen R, Sharmin I, Yusuf MA. Bacteriological Profiles of Pus with Antimicrobial Sensitivity Pattern at a Teaching Hospital in Dhaka City. Bangladesh J Infect Dis. 2018;5(1):10-14.
Crossref - Subha M, Srinivasagam M. Microbial Profile and Antimicrobial Susceptibility Pattern of Pus Culture Isolates from a Teaching Tertiary Care Hospital, South India. Int J Curr Microbiol Appl Sci. 2018;7(4):1149-1153.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.