ISSN: 0973-7510
E-ISSN: 2581-690X
As one of the most common bacterial infections globally, urinary tract infections (UTI)s affect the bladder and kidneys of many the bladders and kidneys of many. Along with gram-negative bacteria, Escherichia coli (E. coli) causes nearly 40% of nosocomial UTIs, 25% of recurrent infections, and between 80 to 90% of community-acquired infections. Proteomics, commonly used to study changes in protein expression of organisms, can be used to explore candidate biomarkers useful for the diagnosis of pathological conditions. Here, protein profiles of samples from patients diagnosed with E. coli-induced UTI were compared to identify distinctive proteins. Extracted proteins from bacteria from patients’ urine samples were separated into excisable spots using 2D-gel electrophoresis. The gels were then analyzed using Progenesis SameSpot software to select uniquely expressed protein spots, excised, and analyzed by LC/MS. The results were then compared against a database of known proteins. We identified two proteins, outer membrane protein A (OmpA) and RNA polymerase-binding transcription factor (DksA), involved in the survival of E. coli in the harsh environment of the host. We suggest their use as a part of a battery of possible biomarkers proteins for E. coli-induced UTI, and suggest that their overexpression is possibly associated with the stage of infection, early or late.
2D gel electrophoresis, Escherichia coli, UTI, OmpA, DksA
As one of the most common global bacterial infections, urinary tract infections (UTI)s affect predominantly the bladder and kidneys of many1. Annually, more than seven million people from around the globe are diagnosed with UTI infections, of which one million people visit the emergency room due to UTI complications. Globally, UTIs put much economic pressure on national health institutions and so must be combatted2. In fact, by 2003, the cost of treating UTI in adult women reached nearly $2 billion annually in the US alone, which includes doctor visits, prescription of antibiotics, and hospitalization3. This number continued to grow and was estimated to be around $2.8 billion due to 400,000 UTI-related hospitalizations by 20114. Generally, women are more likely to be infected with symptomatic UTIs, where one in three will develop at least a UTI once in her lifetime, due to several factors including but not limited to their age and their sexual activity. In addition, female anatomy and low level of estrogen, particularly post menopause, might increase susceptibility to symptomatic UTIs in females5. Moreover, symptomatic UTIs can be more common in individuals with congenital defects of the urinary tract6,7, individuals with urinary obstruction8, and in patients hospitalized for long time5. Behaviors like using birth control such as condoms or spermicidal sprays might be a cofactor in symptomatic UTIs9. On the other hand, asymptomatic UTIs are found in geriatric patients, diabetic patients, spinal cord injury patients, and at a high incidence in catheterized patients and individuals with poor hygiene habits9-12. Asymptomatic infections occur at high frequency in patient with asymptomatic bacteriuria (ABU), which is mainly caused by Escherichia coli (E. coli). However, ABU infections may protect patients from virulent E. coli-induce UTIs and is suggested to reduce the risk of recurrent infections10. In addition, ABU causing strains are unable to express virulence determinants especially surface exposed adhesion molecules13. Therefore, some types of ABU causing strains such as 83972 can be used as prophylactic treatment10.
While gram-negative bacteria are a major cause of UTIs, E. coli is the causative agent of agent of 25% of recurrent UTIs, nearly 40% of nosocomial UTIs, and between 80 to 90% of community-acquired UTIs2,14. Based on the site of infection, E. coli can be classified into intestinal and extraintestinal pathogenic bacteria with several strains that can act as uropathogenic E. coli (UPEC). Inside the host, UPEC becomes an opportunistic intracellular pathogen by releasing virulence factors to establish intracellular bacterial communities that aid in colonizing the urinary tract causing cystitis and pyelonephritis. In response to UPEC colonization, the host’s immunity system is activated and produces cytokines, causing neutrophil influx, and the exfoliation of infected bladder epithelial cells15. UPEC starts to adapt and grow in the new environment by releasing some of its virulence factors coded for by genes of iron acquisition systems, adhesions, and secreted toxins; these genes that are often encoded within regions known as pathogenicity islands16. Consequently, UPEC binds to new cells and replicate within endosome-like structures of the cells from where can cause recurrent UTIs17. A study conducted to showcase genomic differences between ABU strains, UPEC strains (536 and CFT073), and non-pathogenic laboratory strains (MG 1655 (k-12) and Nissle 1917) revealed that differences in virulence genes are due to point mutations, DNA rearrangements, and DNA deletions. The study showed that residual ABU strains were found to have no virulence determinants and were unrelated to UPEC18. Nonetheless, what is of importance is the fact that the same strain isolated from different sites could exhibit alternate virulence factors. For instance, Hly virulence factors were found in many E. coli isolates from women with cystitis and only in a few fecal isolates, and the PapC gene and adhesion genes were found in 41 out of 42 urine isolates but only in 10 fecal ones19. Such studies specifically confirm the differences in proteomic profiles of E. coli isolates taken from urinary tract and those isolated from feces. Smith and colleagues showed proteomic diversities in proteomic profiles among 34 clinical E. coli isolates taken from UTI cases. The researchers employed 2D gel electrophoresis (2DGE), chromatography and mass spectrometry for protein identification and revealed that every patient has his own unique proteome or bacterial population different from the other patient. What is more interesting, researchers found that the proteome of E. coli isolated from a patient with a three-months apart recurrent infection were indistinguishable, and so suggesting that recurrent infection might be caused by the same bacterial population. Similar to other research, they found differences in proteomes of E. coli isolates from urinary tract versus those isolated from feces14.
Proteomics analysis is commonly used to study the differences between virulent and avirulent strains of the same organism. However, it might be challenging as bacterial expressed protein profiles change depending on their growth conditions14. The importance of proteomics lies in its ability to detect post translational modifications of proteins, while the genomics has ability to predict the synthesized proteins20. Similar work using sequential passaging was used to trigger adaptation and alteration of the proteomic profile of E. coli isolated from several patients led to the identification of survival-associated proteins at higher expression, and hence factors aiding in the recurrence of UTI21. Here, the protein profiles of samples from patients diagnosed with E. coli-induced UTI were compared to identify new proteins and add to a database of distinctive ones to be explored in future studies and suggest a fingerprint to rapidly diagnose UTI.
E. coli Isolates
Secondary use of seven anonymous de-identified samples from cases of symptomatic E. coli induced UTIs, originally obtained for medical laboratory testing and not intended for this project, were collected from the Aberdeen Royal Infirmary; ethical committee approval was not necessary. The samples were numbered according to their collection (7, 14, 15, 18, 272, 274, and 277). Samples were streaked for single-colony isolation on MacConkey agar plates and aerobically incubated at 37°C overnight. Isolates suspected to be E. coli were selected and were checked by matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF) biotyper (Bruker Daltonics, Germany) following the manufacturer’s protocols to confirm presence of E. coli.
Microorganism Profiling using MALDI Biotyper
Identification of bacterial isolates were done as previously described with slight modifications22. Briefly, washed E. coli isolates from Colombian agar, fixed in 70% formic acid and acetonitrile, were spread onto a steel MALDI target plate and let to dry. 1 µL of 1% α-cyano-4-hydrocinnamic acid (HCCA) matrix (Bruker, Bremen-Germany) suspension in organic solvent (2.5% trifluoracetic acid and 50% acetonitrile) was overlaid on dry bacterial pellets and left to dry at room temperature. The processed bacterial isolates were tested and analyzed using Bruker Daltonik MALDI-TOF Biotyper (Bruker Daltonik, Germany).
Protein Extraction
Briefly, single colonies were spread on Colombian agar and aerobically incubated overnight at 37 °C. The bacteria was collected from the plate and rinsed in 1 ml of phosphate buffered saline solution (PBS) in microcentrifuge tubes. Next, the tubes were spun down for 5 minutes at 11,000 RPM to discard the supernatant. The pellets were then washed three times in 1 mL of PBS before the final resuspension in 1 mL of the same solution. To determine the accurate mass of bacteria, a 1:50 dilution was prepared using 100 μL of the final bacterial suspension and the optical density (OD) was measured at 600 nm using a spectrophotometer. The remainder of the original bacterial suspension was recentrifuged at 13,000 RPM for 5 minutes, supernatants was completely removed, and pellets were resuspended in 2DGE lysis buffer (Bio-Rad, Germany) at a concentration of 100 µL for every 0.1 OD600. Next, suspensions were placed in ice for 10 minutes, sonicated 3 times at 20-seconds intervals using the Sonic Dismembrator FB-505 (Fisher Scientific, USA), and spun down for 5 minutes at 13,000 RPM. Finally, the supernatants containing bacterial lysate were replaced in newly labelled microcentrifuge tubes and stored at -80°C for later analysis.
Bacterial Proteins Analysis using 2DGE
Two-dimensional gel electrophoresis was carried out following the manufacturer protocol. Briefly, for isoelectric focusing, phase one of the procedure, 125 µL of the lysate was placed in a reswelling tray. Next, 7 cm of immobline pH gradient dehydrated IPG strips (pH range 4-7, GE Healthcare) were placed into the lysate protein samples. A pH range of 4-7 provided adequate resolution as previously published23. The strips were overlaid with Dry Strip Cover Fluid (GE Healthcare, Sweden) and left covered overnight to rehydrate. Then, the now rehydrated IPG strips were electrophoresed in the Multiphor II electrophoresis system (GE Healthcare, Sweden), with the basic ends placed toward the cathode. Two filter wick strips cut to 11 cm were moistened in MilliQ water and placed over the edges of the IPG strips. The electrophoresis protocol was as follows: 1 minute at 200 V, 90 minutes at 3,500 V, then finally 65 minutes at 3500 V; the current was set at 200 Amperes, and the power was set at 5 Watts. Following electrophoresis, strips were removed, blotted then stored at -20°C. Next, the second dimension gel electrophoresis and phase two of the procedure was carried out following the manufacturer protocol. Briefly, after warming up at room temperature, the IPG gel strips were equilibrated with 10 mg/mL dithiothreitol (DTT) in IPG equilibration buffer 34 g of ammonium sulphate, 4 mL of phosphoric acid, 68 mL of methanol in 100 mL of water for 30 minutes while softly rocking side-to-side, then in 25 mg/mL iodoacatamite buffer for 30 minutes while softly rocking side-to-side. Next, IPG gel strips were washed in MilliQ water then in electrophoresis running buffer. The strips were then placed in the designated troph in the Invitrogen precast gels cassett (Bio-Rad, Germany), assembled in the SE250 electrophoresis units (GE Healthcare, Sweden), and electrophoresed for one hour at 75 V then for 2.5 hours at 150 V. Once the run was complete, gels were removed from their plastic casting and fixed while softly rocking side-to-side overnight in a fixative solution (2% mL phosphoric acid and 50% ethanol in MilliQ water). Gels were then washed three times for 30 minutes in MilliQ water then stained while softly shaking with 0.5 g of Coomassie brilliant blue G250 stain in equilibration buffer (Bio-Rad, Germany) for up to 4 days. Finally, after washing gels three times in MilliQ water, the gels were scanned using ImageScanner III (GE Healthcare) to generate 16-bit, 600 dpi, image files for analysis.
Protein Profiles Comparative Analysis
For comparative analysis between samples, the gels scans were analyzed by Progenesis SameSpots software, version 4 (Nonlinear Dynamics, UK). In brief, to eliminate random errors due to loading variation, the software selects a reference gel amongst all sample gels based on a least-significant difference (LSD) post hoc ANOVA analysis of the images at a significance limit of 0.05, hence identifying only statistically unique spots. The Same-Spots software internal algorithms normalize the averages of spot intensities against the selected reference as ratios. To identify large variation possibly caused by genuine up- and down-regulation of proteins are calculated on a log scale above or below a zero average. Each unique spot is then given a number identifiable by the Progenesis SameSpot software. Two protein spots were selected based on their peculiarity or prominent change, and ease of excision from the gel.
Protein Spots Analysis by LC-MS/MS
Protein spots labled by Progenesis SameSpots software to be unique were selected to be identified using LC-MS/MS. Spots were excised based on clarity and ease of excision to avoid contamination from the stained gel, placed in a labelled microcentrifuge tubes and reduced for 20 minutes at 60°C with DTT, S-alkylated for 10 minutes at 25°C with iodoacatamite, and finally digested for 8 hours at 37°C with trypsin. The treated sample was then dried by rotary evaporation, redissolved in 0.1% formic acid, then transferred to a 96-well sealed plate for LC-MS/MS analysis. Finally, using the Mascot program, LC results were analyzed and a score list of best matched proteins is generated based on a non-probabilistic basis for ranking.
Bacterial Strains
The bacterial strains of isolates from the seven samples were verified by MALDI-TOF biotyper then classified using BDMBCR to be one of three strains associated with UTIs (Table 1).
Table (1):
Bacterial strains identified by Bruker Daltonik Biotyper.
Sample |
Bacteria |
Strain |
Score |
|---|---|---|---|
7 |
Escherichia coli |
Escherichia coli DH5alpha BRL |
2.294 |
15 |
Escherichia coli |
Escherichia coli DH5alpha BRL |
2.174 |
18 |
Escherichia coli |
Escherichia coli DH5alpha BRL |
2.145 |
14 |
Escherichia coli |
Escherichia coli MB11464_1 CHB |
2.123 |
274 |
Escherichia coli |
Escherichia coli MB11464_1 CHB |
2.288 |
277 |
Escherichia coli |
Escherichia coli MB11464_1 CHB |
2.131 |
272 |
Escherichia coli |
Escherichia coli B421 UFL |
2.019 |
Protein Identification
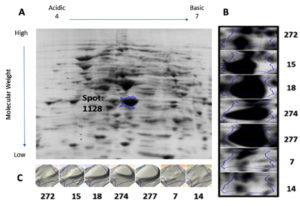
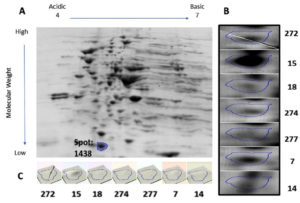
Protein samples extracted from the bacterial cultures isolated from the seven UTI samples were analyzed by 2DGE and Progenesis SameSpots software. The excised spots were then analyzed using LC-MS/MS and the Mascot program to identify the protein. The software aligns the selected images for side-by-side comparison of spots common to all samples (Figure 1 and Figure 2, Panel B). The program also generated a 3D montage from the selected spots of interest from the different gels representing the samples for better visualization of the intensity of the stain, and so the relative amount of expressed protein (Figure 1 and Figure 2, Panel C).
Fig. 1. 2D gel electrophoresis of gel from sample 18 (A). Comparison of spot 1128, identified as OmpA, in all samples (B). 3D montage to visualize the amount of protein in each gel at the same spot (C).
Fig. 2. 2D gel electrophoresis of gel from sample 15 (A). Comparison of spot 1438, identified as DksA, in all samples (B). 3D montage to visualize the amount of protein in each gel at the same spot (C).
Spot number 1128 was highly prominent in all samples except in samples 7 and 14 and was excised from sample number 18 (Figure 1). Spot number 1438 was very weakly observed in samples number 18 and 274, weakly observed in samples 7 and 272, and highly prominent in sample number 15 and accordingly was excised from it
(Figure 2). For spot 1128 from sample number 18, the most significant hit by LC-MS/MS with a Mascot score of 198 was identified as outer membrane protein A (OmpA). As for spot 1438 from sample number 15, the most significant hit by LC-MS/MS with a Mascot score of 737 was identified as RNA polymerase-binding transcription factor DksA.
UTIs are the most common bacterial infection affecting the young and old and should be readily treated to avoid serious complications. Currently, the diagnosis of UTIs is dependent on patient reporting of symptoms and laboratory testing, however the identification of the bacterial agent causing it can be lengthy24. Hence, the search for biomarkers to quickly identify bacterial agent causing illnesses is one of the hallmarks of medical technology and advancement in early diagnosis and treatment. In this study, we aimed to identify key proteins that could be a part of a signature panel battery of proteins suggested to be used as biomarkers in the fast diagnosis of E. coli-induced UTI. Samples were collected at random from seven patients diagnosed with symptomatic E. coli-induced UTIs. The samples were also treated equally under the same conditions to control environmentally driven changes in the protein profile. Nonetheless, much variability in the expressed proteins profile was observed, even among samples identified as the same strain of E. coli (Table 1) and could be attributed to the interaction between the bacteria and the immune system of its host. Furthermore, changes in protein profiles of bacteria may be due to longer incubation in the host and recurrent infections14.
We identified two proteins, outer membrane protein (OmpA) and RNA polymerase-binding transcription factor DksA, taken from two unique spots using LC-MS/MS. Outer membrane proteins (OMPs) are common in gram-negative bacteria and is the major protein of E. coli outer membrane. It is comprised of 3 different domains, a peptidoglycan binding domain, a transmembrane beta-barrel, and a hydrophilic extracellular domain25. OmpA is involved in passive and active transport, catalysis, pathogenesis, and signal transduction in gram-negative proteins26. With a beta-barrel structure, critical for outer membrane stability, OmpA also acts as a channel for passive diffusion25. While Omp proteins are generally involved in adaptation, OmpA, along with other Omp proteins are involved in antibiotic resistance such as tetracycline27. Recent work identified several polymorphisms patterns in extraintestinal pathogenic E. coli including UPEC in the OmpA gene resulting in different protein sequences and thus virulence and response to the host’s defense28. There are several pathways that OmpA, like few other virulence factors, can interact with the host to evade the immune response and grow even in human serum29. For instance, OmpA inhibits the classical pathway of complement activation in E. coli K1 via binding to C4 binding protein (C4BP) to prevent complement-mediated killing30. Furthermore, when OmpA interacts with polymorphonuclear leukocytes, it downregulates several protein-components of NADPH oxidase, a key enzyme responsible for producing the reactive oxygen species important in killing foreign microbes31. Also, OmpA was shown to suppress inflammation and cytokines response by repressing the expression of TNF-α and IL-1β amongst others32. Finally, OmpA has been shown that it aids in the adherence of Acinetobacter baumannii to epithelial cells and biofilm formation33. Likewise, OmpA was shown to be necessary for epithelial cell invasion and the first intracellular replication in the E. coli-induced UTI mouse model34. It is no surprise that Smith et al. dubbed OmpA the “molecular Swiss army knife” of the cell35. Here, we show that OmpA was highly upregulated in five of the seven samples tested in this study, such upregulation is a result of adaptation or survivor mechanism to combat the host’s immune system. However, the variability of the expression amongst the samples needs further investigation. Although such variability could be due to several factors, one of which is the stage of the infection, we hypothesize that samples showing the upregulated OmpA was at a peaking stage of the infection as opposed to the samples with no OmpA expression that could be at a starting stage of it.
Our investigation also identified a second possible candidate that could be part of a protein battery to identify and stage E. coli-induced UTIs, the RNA polymerase-binding transcription factor, DksA. This protein directly binds to RNA polymerase (RNAP) and alters transcription, regulation of a number of amino acid biosynthesis, and their rRNA expression36. Binding of DksA to RNAP interferes with RNAP and DNA interaction, inhibits initiation complexes formation, and amplifies the effects of NTP and guanosine-3′, 5′-bis(diphosphate) (ppGpp) on rRNA transcription37. Although DksA is involved in many processes in E. coli, it is worthy to mention that it plays a role in the rigorous metabolic response to environmental stressors such as those inflected by the host immune system38,39. Along with ppGpp, DksA is a key element in the stringent response that represses rapid growth genes, activates survival genes, and synthesizes amino acids40. In fact, when DksA is either deleted or overexpressed, its pleiotropic nature caused defects in gene expression, cell division, and amino acid biosynthesis amongst other factors considered virulent37.
Taking it all, our research showed that both OmpA and DksA were strongly upregulated in only one sample, number 15, as opposed to other samples that exhibited the upregulation of OmpA and only a slight upregulation of DksA, if any. Again, we hypothesize that while OmpA plays a role in the early stages of infection leading to its peak such as adhering to epithelial cells, resisting compliments in the serum, and combating PMNs, DksA plays a role in a harsher environment requiring growth reduction and metabolism reprogramming, necessary for survival in a later ending stage of the infection. Furthermore, this suggests that OmpA could be associated with acute cases, while DksA could be associated with chronic ones. Unfortunately, since the samples were from a bank of secondary-use samples not originally intended for this preliminary investigation, no information can be retrieved regarding the individual cases to be related to such suggestions. Therefore, we recommend that our findings should only be considered as a pilot study requiring further correlation with patient history and confirmation with larger sample size. We suggest that combined with our recently published work that identified four protein candidates specific to E. coli, ASPA ECOLI, ATPB_ECOBW, DCEB ECOLI, and DPS ECOL6 21, the two proteins identified here can also be candidate to stage the infection, early or late.
ACKNOWLEDGMENTS
The authors would like to acknowledge King Abdulaziz City for Science and Technology (KACST) for its support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AS completed the experimental work and wrote the initial manuscript, GA provided the tested concept and reviewed the manuscript, AM supervised the work, reviewed and edited the manuscript.
FUNDING
King Abdulaziz City for Science and Technology (KACST) grant No. 1-17-00-009-0005
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Barber AE, Norton JP, Spivak AM, Mulvey MA. Urinary tract infections: current and emerging management strategies. Clin Infect Dis. 2013;57(5):719-724.
Crossref - Cai W, Wannemuehler Y, Dell’anna G, et al. A novel two-component signaling system facilitates uropathogenic Escherichia coli’s ability to exploit abundant host metabolites. PLoS Pathog. 2013;9(6):e1003428.
Crossref - Rosenberg M. Pharmacoeconomics of treating uncomplicated urinary tract infections. Int J Antimicrob Agents. 1999;11:247-251.
Crossref - Simmering JE, Tang F, Cavanaugh JE, Polgreen LA, Polgreen PM. The Increase in Hospitalizations for Urinary Tract Infections and the Associated Costs in the United States 1998-2011. Open Forum Infect Dis. 2017;4(1).
Crossref - Foxman B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Disease-a-Month. 2003;49(2):53-70.
Crossref - Sheinfeld J, Schaeffer AJ, Cordon-Cardo C, Rogatko A, Fair WR. Association of the Lewis Blood-Group Phenotype with Recurrent Urinary Tract Infections in Women. N Engl J Med. 1989;320(12):773-777.
Crossref - Twaij M. Urinary tract infection in children: A review of its pathogenesis and risk factors. J R Soc Promot Health. 2000;120(4):220-226.
Crossref - El-Dahr SS, Lewy JE. Urinary tract obstruction and infection in the neonate. Clin Perinatol. 1992;19(1):213-222.
Crossref - Hooton TM, Scholes D, Hughes JP, et al. A Prospective Study of Risk Factors for Symptomatic Urinary Tract Infection in Young Women. N Engl J Med. 1996;335(7):468-474.
Crossref - Sunden F, Hakansson L, Ljunggren E, Wullt B. Escherichia coli 83972 Bacteriuria Protects Against Recurrent Lower Urinary Tract Infections in Patients With Incomplete Bladder Emptying. J Urol. 2010;184(1):179-185.
Crossref - Gonzalez CM, Schaeffer AJ. Treatment of urinary tract infection: what’s old, what’s new, and what works. World J Urol. 1999;17(6):372-382.
Crossref - Stamm WE, Hooton TM. Management of Urinary Tract Infections in Adults. N Engl J Med. 1993;329(18):1328-1334.
Crossref - Imirzalioglu C, Hain T, Chakraborty T, Domann E. Hidden pathogens uncovered: Metagenomic analysis of urinary tract infections. Andrologia. 2008:40;66-71.
Crossref - Smith A, van Rooyen JP, Argo E, Cash P. Proteomic analysis of Escherichia coli associated with urinary tract infections. Proteomics. 2011;11(11):2283-2293.
Crossref - Bower JM, Eto DS, Mulvey MA. Covert operations of uropathogenic Escherichia coli within the Urinary Tract. Traffic. 2005;6(1):18-31.
Crossref - Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85(1):11-19.
Crossref - Mysorekar IU, Hultgren SJ. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc Natl Acad Sci USA. 2006;103(38):14170-14175.
Crossref - Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U. Molecular basis of commensalism in the urinary tract: Low virulence or virulence attenuation? Infect Immun. 2008;76(2):695-703.
Crossref - Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol. 2008;46(8):2529-2534.
Crossref - Cash P. Proteomics of bacterial pathogens. Expert Opin Drug Discov. 2008;3(5):461-473.
Crossref - Alhajouj MS, Alsharif GS, Mirza AA. Impact of Sequential Passaging on Protein Expression of E. coli Using Proteomics Analysis. Int J Microbiol. 2020;2020:2716202.
Crossref - Seng P, Drancourt M, Gouriet F, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49(4):543-551.
Crossref - Park MR, Lee EG, Kim YH, et al. Reference map of soluble proteins from Salmonella enterica serovar Enteritidis by two-dimensional electrophoresis. J Vet Sci (Suwon-si, Korea). 2003;4(2):143-149.
Crossref - Masajtis-Zagajewska A, Nowicki M. New markers of urinary tract infection. Clin Chim Acta. 2017;471:286-291.
Crossref - Wang Y. The function of OmpA in Escherichia coli. Biochem Biophys Res Commun. 2002;292(2):396-401.
Crossref - Alteri CJ, Mobley HLT. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect Immun. 2007;75(6):2679-2688.
Crossref - Viveiros M, Dupont M, Rodrigues L, et al. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS One. 2007;2(4):365.
Crossref - Nielsen DW, Ricker N, Barbieri NL, Allen HK, Nolan LK, Logue CM. Outer membrane protein A (OmpA) of extraintestinal pathogenic Escherichia coli. BMC Res Notes. 2020;13:51.
Crossref - Rooijakkers SHM, van Strijp JAG. Bacterial complement evasion. Mol Immunol. 2007;44(1-3):23-32.
Crossref - Prasadarao NV., Blom AM, Villoutreix BO, Linsangan LC. A Novel Interaction of Outer Membrane Protein A with C4b Binding Protein Mediates Serum Resistance of Escherichia coli K1 . J Immunol. 2002;169(11):6352-6360.
Crossref - Krishnan S, Prasadarao NV. Outer membrane protein A and OprF: Versatile roles in Gram-negative bacterial infections. FEBS J. 2012;279(6):919-931.
Crossref - Selvaraj SK, Prasadarao NV. Escherichia coli K1 inhibits proinflammatory cytokine induction in monocytes by preventing NF-κB activation. J Leukoc Biol. 2005;78(2):544-554.
Crossref - Kim SW, Choi CH, Moon DC, et al. Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol Lett. 2009;301(2):224-231.
Crossref - Nicholson TF, Watts KM, Hunstad DA. OmpA of uropathogenic Escherichia coli promotes postinvasion pathogenesis of cystitis. Infect Immun. 2009;77(12):5245-5251.
Crossref - Smith SGJ, Mahon V, Lambert MA, Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett. . 2007;273(1):1-11.
Crossref - Parshin A, Shiver AL, Lee J, et al. DksA regulates RNA polymerase in Escherichia coli through a network of interactions in the secondary channel that includes Sequence Insertion 1. Proc Natl Acad Sci USA. 2015;112(50):E6862-E6871.
Crossref - Paul BJ, Barker MM, Ross W, et al. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118(3):311-322.
Crossref - Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: The intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol. 2015;197(7):1146-1156.
Crossref - Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13(5):298-309.
Crossref - Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102(22):7823-7828.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.