ISSN: 0973-7510
E-ISSN: 2581-690X
About 20 % of neonates develop sepsis and among them approximately 1% die due to sepsis-related causes. Bacterial pathogens are the commonest cause of neonatal sepsis which is either early-onset (<72 hours of age) or late-onset (>72 hours). Little is known about the epidemiology and antimicrobial susceptibility pattern of sepsis causing bacterial pathogens in Nepal. A prospective study was carried out among neonates suspected to have sepsis and admitted to Tribhuwan University Teaching Hospital from January to December 2016. Clinical suspicion of sepsis was made based on clinical findings and laboratory parameters, later confirmed by isolation of organisms in blood culture. Drug resistance pattern of Gram-positive and Gram-negative bacteria were studied by standard methods. Meropenem resistant Gram-negative bacteria were processed for the detection of β-lactamases and resistant genes were detected by X-pert Carba-R (Cepheid) Assays. Of 372 neonates with clinically suspected sepsis, 132 (35.4%) had blood culture positivity, with 47% early-onset and 53% late-onset sepsis. Coagulase-negative Staphylococcus aureus (CONS) was the most common (37.9%) etiological agent followed by Klebsiella pneumoniae (12.9%). Of all 132 isolates, 81 (61.3%) were Gram-positive of which 22 (27.2%) were multi-drug resistant (MDR), three (3.7%) were methicillin-resistant S. aureus (MRSA), and 14 (17.2%) were methicillin-resistant CoNS; and 50 (37.8%) were Gram-negative of which 26 (52%) were MDR and 29 (58%) were resistant to β-lactamases. The blaKPC gene was detected in four isolates of K. pneumoniae, two of E. coli, one ABC (Acinetobacter baumanii complex), and one Enterobacter aerogenes whereas blaNDM gene was detected in one isolate of K. pneumoniae, two of E. coli, two Pseudomonas aeruginosa, one Acinetobacter baumanii complex, and one Enterobacter aerogenes. Overall mortality due to sepsis-related causes was 7.6% (10 of 132). One-third of clinically suspected neonatal sepsis cases were culture positive. Late-onset sepsis was more common than early onset. CoNS was the predominant bacterial isolate followed by Klebsiella pneumoniae, with high rates of multi-drug resistance.
Neonatal Sepsis, MDR, Nepal
Neonatal sepsis (NS) is a clinical syndrome presented with clinical features of infection with or without underlying bacteremia in the first month of life.5 It is a leading cause of neonatal morbidity and mortality worldwide.1 About 30-50% of total neonatal deaths are due to NS in developing countries.2,3 According to the World Health Organization (WHO) reports, about 20% of all neonates develop sepsis and approximately 1% die due to sepsis-related causes.3 Those who survive are prone to short and long-term morbidity (neurodevelopmental).4 Sepsis-related morbidity and mortality is largely preventable with the prevention of sepsis itself, with timely recognition of the disease and by appropriate and aggressive antimicrobial therapy and supportive care.
Neonatal sepsis (NS) is classified as early onset and late onset sepsis.5
Early onset sepsis (EOS): Presentation within the first 72 hours of life. Infants with EOS mostly present with pneumonia and respiratory distress. Maternal genital tract is the primary source of infection.5 An increased risk of EOS is seen with some maternal-perinatal conditions.5 Early diagnosis of sepsis can be done with the knowledge about these potential risk factors.5
Based on the studies in India, an increased risk of early onset sepsis has been seen with the following risk factors: (a) Febrile illness in the mother within two weeks prior to delivery with evidence of bacterial infection; (b) Prematurity or Low birth weight (<2500 grams); (c) Rupture of membranes >24 hours; (d) Single unclean or >3 sterile vaginal examinations during labor; (e) Prolonged labor (sum of first and second stage of labor >24 hours); (f) Foul smelling and/or meconium stained liquor; (g) Perinatal asphyxia (Apgar score <4 at one minute).5
Antibiotic treatment should be initiated for the newborn if there is foul-smelling liquor or three of the above-mentioned risk factors. If only two risk factors are present then infants should be investigated for possible infection and then treated accordingly.5
Late onset sepsis (LOS)
Presentation after 72 hours of life. Nosocomial (hospital-acquired) or community-acquired can be source of infection in LOS.5 Neonates’ usual presentation are with pneumonia, septicemia or meningitis. Predisposing factors that increased risk of nosocomial sepsis include prematurity, low birth weight, invasive procedures, mechanical ventilation, admission in intensive care unit, administration of parenteral fluids, and use of stock solutions.5
Neonatal sepsis can be classified into three major groups according to the criteria for the diagnosis of neonatal sepsis.6
- Proven sepsis: Infants with clinical signs and symptoms of infection plus a positive blood culture or PCR. For coagulase-negative Staphylococcus aureus (CoNS) either two positive blood cultures are required or one positive blood culture plus an increase in inflammatory markers like positive (i.e. >6 mg/dL) C-reactive protein (CRP).6
- Probable Sepsis: When blood culture is negative, presence of clinical signs and symptoms of infection with at least two abnormal laboratory results are required.6
- Possible Sepsis: When blood culture is negative, presence of clinical signs and symptoms of infection plus raised in inflammatory markers like CRP or IL-6/IL-8 level.6
The organisms that cause EOS of term and preterm infants together are Group B Streptococcus (GBS) and E. coli, which is responsible for approximately 70% of infections combined.5,6 Intrapartum antibiotic prophylaxis which has decreased dramatically the incidence of early-onset GBS infection but it still remains a major cause of neonatal sepsis. Others organism that cause EOS are Streptococcus viridans, Streptococcus pneumoniae, Enterococcus spp., Staphylococcus aureus and Gram-negative enteric bacteria such as Enterobacter spp., Haemophilus influenza (virtually all non-typeable Haemophilus spp.), and Listeria monocytogenes.6 The organisms associated with LOS are CoNS, S. aureus, E. coli, K. pneumoniae, Enterobacter spp., Pseudomonas aeruginosa, and Acinetobacter spp.7
Neonatal sepsis continues to remain a common and significant health care burden in low-resource settings, especially in vulnerable groups such as very-low-birth-weight infants (VLBW:<1500 grams). Despite ongoing efforts for early diagnosis, treatment, and prevention, it remains a challenge for clinicians due to frequent changes in epidemiology, non-specific symptoms, lack of ideal diagnostic markers, and emergence of multi-drug resistant (MDR) pathogens.8,9
This study was conducted among neonates, both in-born and brought-in cases, admitted to the Neonatal Intensive Care Unit (NICU) and non-NICU units (neonatal unit and neonatal ward) at the Department of Pediatrics in Tribhuwan University Teaching Hospital (TUTH) from January to December 2016. All newborns with clinical suspicion of sepsis were enrolled into the study and their body samples (blood, urine, cerebrospinal fluid, pus, sputum) were processed for sepsis work-up. In this study, blood cultures were only analyzed because blood culture positivity is a prerequisite for labelling any case a ‘proven sepsis’.6 Clinical suspicion of sepsis was made on the basis of following variables.6,10
Clinical variables – a) Temperature instability; b) tachypnea plus grunting or desaturations; c) tachycardia or bradycardia; d) plasma glucose >10 mmol/L; e) feeding intolerance; f) lethargy/altered mental status.
Hemodynamic variables – a) Blood pressure two Standard Deviation below normal for age (Systolic pressure <50 mmHg for newborn on Day-1, <65 mmHg for infants <1 month).
Tissue perfusion variables – a) Capillary refill time >3 seconds; b) plasma lactate >3 mmol/L.
In clinically suspected cases, additionally, the following inflammatory variables were investigated. Positivity for any two of them would make the case ‘probable sepsis’ in absence of blood culture positivity.
- Semi-quantitative CRP test performed by Latex agglutination method. CRP level >6 mg/dL was considered positive.
- Total leukocyte count <5,000 or >30,000 cells per mm.3
- Micro-ESR >15 mm in first hour
- Platelet count <100,000 cells/μL
Micro-ESR was performed in the bed side; total leucocyte count (TLC), differential count (DC), and platelet count were tested in the hematology laboratory; and blood culture and C-reactive protein (CRP) tests in microbiology laboratory by following the standard methods.11
One ml of blood was mixed with nine ml of Brain Heart Infusion (BHI) broth under aseptic condition then transported to the microbiology laboratory. BHI broth with blood samples were incubated at 37°C aerobically and observed for turbidity and other signs of bacterial growth. Sub-culture was done in Chocolate, Blood and MacConkey agar plates from turbid BHI broths and incubated at 37°C for 16-18 hours. The organisms were isolated and identified as Gram-positive or Gram-negative according to the American Society for Microbiology (ASM) guidelines.11
Results of Gram staining test were informed to the clinicians. Antibiotic susceptibility testing (AST) was done simultaneously with isolate identification and reports were dispatched accordingly. Patterns of MDR (i.e. organism is resistant to at least three groups of antimicrobial agents) were also noted.
In Gram-positive organisms, methicillin resistance was identified according to Clinical and Laboratory Standard Institute (CLSI) guidelines. In Gram-negative cases, cefotaxime resistant organisms were tested for Extended Spectrum β-lactamase (ESBL) and AmpC type β-lactamase.
Modified Hodge test was performed in meropenem resistant organisms following the CLSI guidelines. Organisms that tested positive for Modified Hodge test were processed for the detection of Metallo-β-lactamase (MBL), Klebsiella pneumonia carbapenemase type β-lactamases (KPCs), and Oxacillinase type β-lactamases (OXA) drug resistance patterns according to the standard guidelines.7,9 For genetic confirmation of drug resistance mechanism, all of the isolates were processed for X-pert Carba-R (Cepheid) Assay. (Figures 1-10)
Fig. 1. 9ml of BHI broth from left to right showing only BHI broth, BHI broth with 1ml of blood but without turbidity and BHI broth with 1ml of neonatal blood with turbidity.
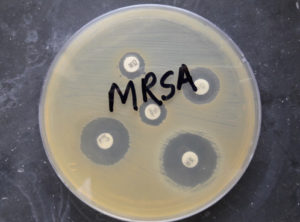
Fig. 4. Methicillin-resistant Staphylococcus aureus (MRSA) showing resistant to cefoxitin 30 microgram
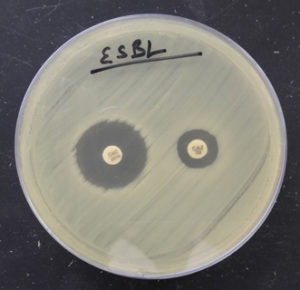
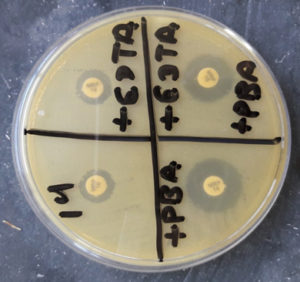
Fig. 5. Organism producing ESBL detected by combined disk method (zone size increased by >5mm in combination disk of ceftazidime plus clavulanic acid in comparison to ceftazidime alone).
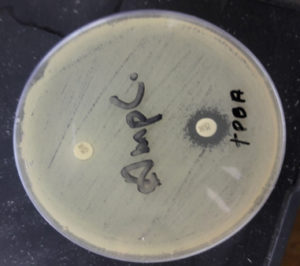
Fig. 6. Organism producing AmpC detected by cefoxitin (CX 30) and cefoxitin (CX 30) plus phenyl boronic acid (zone size increased by >5mm in CX 30 plus phenylboronic acid in comparison to CX 30 alone).
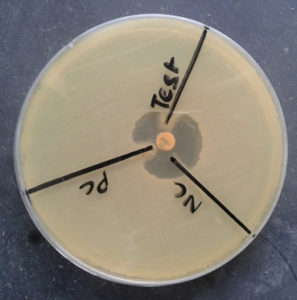
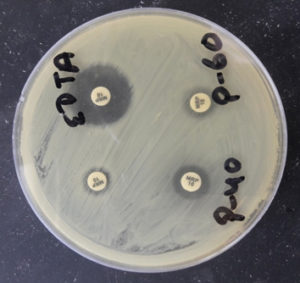
Fig. 8. Detection of MBL by using Meropenem and EDTA (Meropenem plus EDTA disk showing increase in zone diameter by >5mm in comparison to Meropenem disk only).
Fig. 9. Detection of KPCs by using Meropenem and phenyl boronic acid (Meropenem plus phenyl boronic acid disk showing increase in zone diameter by >5mm in comparison to Meropenem disk only).
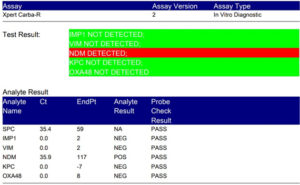
Fig. 10. Detection of NDM by using Xpert Carba-R Assay (Genetic confirmation by detecting blaNDM drug resistance gene).
The patients included into the study were prospectively followed up until discharge or death. All reports were recorded in the case report forms (Performa).
IRB of Institute of Medicine, TUTH has approved this study. Informed written consent was taken from each patient’s parent or guardian before enrollment into the study. Descriptive analysis of data was done by SPSS Inc. version 21.0.12 Results were interpreted as frequency distribution and percentages.
During one-year study period, 372 neonates were admitted to the neonatal units with the diagnosis of suspected sepsis, 59.1% were male and 40.9% were female; (52.4%) were <72 hours age and (47.6%) were >72 hours age.
Of 372 suspected sepsis cases, 132 (35.5%) had a positive and 240 (64.5%) had a negative blood culture. Nearly 47% of culture-positive cases fell into early-onset category and 53% (70 of 132) into late-onset sepsis category.
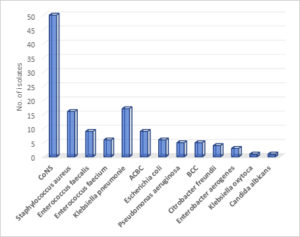
Overall, the most common isolates were coagulase-negative Staphylococcus (CoNS) (37.9%, 50 of 132) followed by Klebsiella pneumoniae (12.9%, 17 of 132). Fig. 1. In EOS, CoNS and Klebsiella pneumoniae were most common isolates, whereas Staphylococcus aureus and Enterococcus species were most common in LOS. Table 1.
Table (1):
Age-wise distribution of blood culture isolates in 132 culture-positive cases.
| Culture isolates | >72 hours (LOS) | < 72 hours (EOS) | Total | |
|---|---|---|---|---|
| No. (%) | No. (%) | No. | % | |
| CoNS | 22 (31·4%) | 28 (45·2%) | 50 | 37·9 % |
| Staphylococcus aureus | 12 (17·1%) | 4 (6·5%) | 16 | 12·1% |
| Enterococcus fecalis | 6 (8·6%) | 3 (4·8%) | 9 | 6·8% |
| Enterococcus faecium | 5 (7·1%) | 1 (1·6%) | 6 | 4·5% |
| Escherichia coli | 4 (5·7%) | 2 (3·2%) | 6 | 4·5% |
| Klebsiella pneumoniae | 8 (11·4%) | 9 (14·5%) | 17 | 12·9% |
| Klebsiella oxytoca | 0 | 1 (1·6%) | 1 | 0·8% |
| BCC | 3 (4·3%) | 2 (3·2%) | 5 | 3·8% |
| ACBC | 5 (7·1%) | 4 (6·5%) | 9 | 6·8% |
| Citrobacter freundii | 2 (2·9%) | 2 (3·2%) | 4 | 3% |
| Enterobacter aerogenes | 1 (1·4%) | 2 (3·2%) | 3 | 2·3% |
| Pseudomonas aeruginosa | 2 (2·9%) | 3 (4·8%) | 5 | 3·8% |
| Candida albicans | 0 | 1 (1·6%) | 1 | 0·8% |
CoNS: Coagulase-negative Staphyloccocus aureus; BCC: Burkholderia cepacia complex; ACBC: Acinetobacter calcoaceticus baumanii complex
Following treatment, 92.4% (122) of culture-positive neonates got improved and discharged from the hospital, whereas 7.6% (10) got expired. Of 122 improved cases, 56 (45.9%) were EOS and 66 (54.1%) were LOS; of 10 expired cases, 6 (60%) were EOS and four (40%) were LOS.
Of all culture-positive sepsis cases, 33 (25%) were preterm, five (3.8%) were low birth weight, six (4.5%) had perinatal asphyxia, and five (3.8%) were preterm with premature rupture of membranes (PROM). Nearly half (48.5%) of the neonatal sepsis cases had no risk factors of neonatal sepsis. Table 2.
Table (2):
Risk factors of neonatal sepsis in 132 culture-positive cases.
Risk factors |
Frequency (%) |
|---|---|
No risk factor |
64 (48.5%) |
Pre-term |
33 (25.0%) |
Perinatal asphyxia |
6 (4.5%) |
Pre-term with premature rupture of membranes (PROM) |
5 (3.8%) |
Low birth weight (LBW) |
5 (3.8%) |
Congenital anomaly with surgical intervention |
4 (3.0%) |
Congenital anomaly |
4 (3.0%) |
Meconium stained liquor |
4 (3.0%) |
LBW with mechanical ventilation |
2 (1.5%) |
Other interventions |
2 (1.5%) |
PROM with perinatal asphyxia |
1 (0.8%) |
Mechanical ventilation |
1 (0.8%) |
PROM |
1 (0.8%) |
As high as 36.6% of bacterial organisms causing neonatal sepsis were found to be multi-drug resistant (MDR): 16.8% (22/131) of Gram positive and 19.84% (26/131) of Gram negative bacteria isolated. Table 3.
Table (3):
Proportion of multi-drug resistant organisms among 131 blood culture isolates.
Bacterial isolates |
Total |
MDR isolates (%) |
|---|---|---|
Gram-positive organisms |
81 |
22 (16.8%) |
Gram-negative organisms |
50 |
26 (19.84%) |
Total |
131 |
48 (36·6%) |
Note: One of the isolates of blood culture was Candida albicans
Among 16 Staphylococcus aureus isolates, three (18.7%) were methicillin resistant (MRSA) and among 50 CoNS isolates, 14 (18%) were methicillin resistant. Out of 50 Gram negative isolates of blood culture, eight (16%) were KPCs; ESBL, AmpC, and MBL were seven (14%) each. Xpert Carba-R Assays confirmed KPCs and New Delhi Metallo-β-lactamase (NDM) by detecting blaKPC and blaNDM drug resistance genes respectively. The blaKPC gene was detected in four isolates of K. pneumoniae, two of E. coli, one ABC, and one Enterobacter aerogenes. blaNDM gene was detected in one isolate of K. pneumoniae, two isolates of E. coli, two isolates of Pseudomonas aeruginosa, one isolates of Acinetobacter baumanii complex (ABC), and one isolates of Enterobacter aerogenes.
Neonatal sepsis continues to remain a major clinical problem with high rates of morbidity and mortality, especially in resource-poor countries like Nepal. In our study, more than one third (35.4%) of neonates had microbiologically confirmed sepsis. This finding is in agreement with other studies done in Nepal in which rates of culture-proven sepsis were 30.8% and 28.3%.7,13
The clinical signs and symptoms in neonates are rather subtle and nonspecific which makes it difficult for an early diagnosis of neonatal sepsis. Sepsis diagnosis may also be interfered by other life-threatening diseases in newborns such as perinatal asphyxia and necrotizing enterocolitis.14,15 Blood culture is still the gold standard test for the definitive diagnosis of neonatal sepsis despite its drawbacks such as low sensitivity, time-consuming, and possible contamination especially with commensal CoNS.4
The rate of culture-proven neonatal sepsis (35.5%) is comparable to those reported in other developing countries of Asia and Africa: Nigeria (45.9%), Ethiopia (44.7%), Bangladesh (34.9%) and Uganda (37%)16-19 However, negative blood culture does not rule out sepsis because about 26% of all neonatal sepsis could be due to anaerobic organism.20 Other pathogens potentially causing sepsis such as viruses (for e.g. rubella, cytomegalovirus), protozoa (for e.g. Toxoplasma gondii), and treponema (for e.g. Treponema pallidum).4
The incidence of septicemia has been reported to be higher in males than in females.21 In our study too, septicemia was found to be higher in males (59.1%) compared to females (40.9%), with male-to-female ratio of 1.4:1. In another study from Nepal, male-to-female ratio was 1.2:1 in 2015 and 2:1 in 2003.7,13. Prevalence of gender bias in the society (especially while seeking health care), place of study, and sample-related factors could be the potential reasons for male preponderance.7
The proportion of late onset sepsis was higher than early onset sepsis in our setting (incidence of LOS vs. EOS: 53% and 47%, respectively). This is in agreement with previous studies.22,23 Nagaveni et al., were reported that LOS and EOS were 53.3% and 46.7%, respectively2, which is consistent with our results In contrast, another study from India by Varsha et al. reported higher rate of EOS than LOS, which could be justified by a larger cohort of neonates in <72 hours age group (74.6%) compared to >72 hours age group (25.3%).23
The most common organism isolated from infected neonates was CONS (37.9%) followed by Klebsiella pneumoniae (12.9%). This finding is in agreement with studies done in India and Egypt which showed CONS as the predominant isolate in both EOS and LOS.21,4 Contrary to our findings of CoNS and Klebsiella pneumoniae as the most common isolates in EOS, a six-year analysis of neonatal sepsis cases in Taiwan have reported Group B Streptococci (GBS) (36%) and Escherichia coli (26%). Similarly, the most common organisms in LOS were Staphylococcus aureus and Enterococcus spp. whereas CoNS (40%) and Candida (15%) were most common in Taiwan.24
Prematurity, low birth weight, and perinatal asphyxia were reported in neonates with culture-positive sepsis. High mortality rates have been reported in neonates with these factors, especially in resource-poor settings.25
Sepsis is a leading cause of neonatal morbidity and mortality. In developing countries it accounts for 30-50% of total neonatal deaths.2,3 It is estimated that about 1% of all neonates die due to sepsis-related causes.3 In our study, neonatal mortality rate in probable sepsis cases was 7.6 % (10/132), of which 60% were EOS and 40% were LOS. The mortality rate in proven sepsis cases was 8.3% (10/120). These findings are in agreement with a study done in India by Nagaveni et al. which reported neonatal mortality rate of 14.6%, of which 56.3% had EOS and 43.7% had LOS.21
It was found that 90.9% (120 of 132) of culture-positive cases in this study were also positive for at least two of five inflammatory laboratory parameters—i.e. proven sepsis. Remaining 12 cases did not fulfil the criteria of sepsis although they had bacteremia. The lab parameters tested were: Total leucocyte count (TLC), platelet count, C-reactive protein (CRP), and micro-ESR. In a study done by Shah B. et al., sensitivity and specificity for TLC, platelet count, and CRP was found to be 44% and 92%, 22% and 99%, and 70-93% and 78-94% respectively. Shah B. et al. used CRP, procalcitonin, Interleukin-6 (IL-6), IL-8, tumor necrosis factor (TNF) alpha, and cell surface markers such as Cd11Beta, CD64 as the biomarkers of sepsis.8
Out of 132 blood culture isolates in our study, 81 were Gram positive, 50 were Gram negative, and one was Candida albicans. Among Gram positives, CoNS, Staphylococcus aureus, Enterococcus faecalis and Enterococcus faecium were isolated, of which 22 (27.2%) were MDR. Of 16 Staphylococcus aureus isolates, 3 (18.7%) were MRSA and out of 50 CoNS isolates, 14 (18%) were methicillin resistant. In proven sepsis cases with Gram positive isolates, vancomycin and teicoplanin are the most effective drug of choice.
Among 50 Gram negative isolates of blood culture, 26 (52%) were MDR, eight (16%) were KPCs, and the rest were ESBL, AmpC, and MBL at 14% (seven each). The Gram-negative isolates were Esherichia coli, Klebsiella pneumonia, Klebsiella oxytoca, Burkholderia cepacia complex, Acinetobacter calcoaceticus baumanii complex, Citrobacter freundii, Enterobacter aerogenes, and Pseudomonas aeruginosa.
Our study found a high rate of antimicrobial resistant bacterial isolates causing neonatal sepsis. The burden of multidrug-resistant organisms is rapidly emerging and it could be potentially disastrous in many settings like ours. Infection with such organisms could lead to treatment failure, higher morbidity and mortality rates, increased health costs, and prolonged hospitalization of patients.7,26
There are few limitations of the study. Blood samples were processed aerobically only, therefore, anaerobic and fastidious slow growing organisms were not grown. Only the phenotypic drug resistance mechanism was studied, so the genetic makeup of the resistant bacteria could not be established. Additionally, for sepsis screening, only four inflammatory markers were included out of eight recommended markers (variables like immature neutrophil, immature: total neutrophil ratio, pro-calcitonin, IL-6 or IL-8, and 16s PCR were not included). The inclusion of all markers could help in detecting sepsis even with blood culture negativity.
In this study, just above one-third of clinically suspected sepsis cases were culture positive and CoNS was the predominant isolate followed by Klebsiella pneumoniae. Rate of multi-drug resistance was also high. Because clinical signs and symptoms are subtle in neonates, a clinical diagnosis of sepsis can be the challenge. Therefore, routine blood cultures along with early information about antibiotic susceptibility and multi-drug resistance patterns could supplement clinical guidance to effective treatment.
Summary of findings
- Out of 372 suspected sepsis cases, 32.3% (120 of 372) were proven sepsis.
- Blood culture positivity was higher among neonates admitted to NICU (43.8%) in comparison to non-NICU admissions (31.9%)
- The most common organism grown in blood culture was CoNS followed by Klebsiella pneumoniae and Staphylococcus aureus.
- LOS was more common than EOS (53% vs. 47%).
- Staphylococcus aureus was more commonly isolated in LOS than in EOS.
- MDR rate in the bacterial isolates of blood culture was 36.6% (48 of 131).
- Along with MDR pattern, ESBL, AmpC, KPCs, and MBL were also detected in Gram negative bacteria, and MRSA and methicillin resistant CoNS in Gram positive bacteria.
- Pre-term birth (25%), perinatal asphyxia (4.5%), low birth weight (3.8%) and PROM (3.8%) were reported in neonates with culture-positive sepsis.
- Mortality rate among blood culture positive neonatal sepsis cases was 7.6% (10 of 132).
Research in context
Evidence before this study
NS is one of the leading causes of neonatal mortality and morbidity worldwide. In developing countries, it is responsible for 30-50% of total neonatal deaths. According to the WHO reports, about 20% of all neonates develop sepsis and among them approximately 1% die due to sepsis-related causes. No uniform data is available in Indian subcontinent including Nepal in relation to etiological agents of NS, mortality trend due to infection, and antimicrobial susceptibility pattern of bacterial isolates. Advanced microbiological tests are rarely available in low-resource countries such as Nepal.
Added-value of this study
This study has assessed the prevalence of culture-proven sepsis among all neonates admitted to the largest tertiary hospital of Nepal (TUTH, Kathmandu) over a one-year study period. Additionally, this study has explored the etiological agents of NS, and tracked the trend of antimicrobial susceptibly and multi-drug resistance. Drug-resistant genes such as blaKPC and blaNDM have been detected in Gram-negative bacterial isolates.
Implications of all the available evidence
Because clinical signs and symptoms are subtle in neonates, their clinical diagnosis of sepsis can be the challenge. Our study findings could be a reference for clinicians in resource-limited settings, who would be encouraged to order routine blood cultures in suspected sepsis cases. Additionally, it is important to get information about antibiotic susceptibility and multi-drug resistance patterns of sepsis-causing bacterial agents early on during case management.
ACKNOWLEDGMENTS
We would like to thank all patients and their parents or guardians for their cooperation and for providing us consent to conduct this study. We would also like to thank all members of the Microbiology and Pediatrics department of Tribhuvan University Teaching Hospital for their help and cooperation throughout the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RS collected data and processed lab reports. NPS, BMP, BR, and SB supervised the study. RS and SB interpreted the data. SB, SS and SS prepared manuscript drafts and reviewed comments. All authors have read and approved the final manuscript.
FUNDING
None.
ETHICS STATEMENT
Ethical approval was taken from the Institutional Review Committee (IRC), Institute of Medicine. Tribhuvan University Teaching Hospital, Kathmandu, Nepal.
AVAILABILITY OF DATA
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
INFORMED CONSENT
Informed consent was taken from the legal guardian of the neonates.
- Liu L, Johnson HL, Cousens S, et al. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379(9832):P2151-P2161.
Crossref - Bang AT, Bang RA, Baitule SB, Reddy MH, Deshmukh MD. Effect of home-based neonatal care and management of sepsis on neonatal mortality: Field trial in rural India. Lancet. 1999;354(9194):P1955-1961.
Crossref - Stoll BJ. The global impact of neonatal infection. Clin Perinatol. 1997;24(1):1-21.
Crossref - Shehab El-Din EMR, El-Sokkary MMA, Bassiouny MR, Hassan R. Epidemiology of neonatal sepsis and implicated pathogens: A Study from Egypt. Biomed Res Int. 2015;2015:509484.
Crossref - Singh M, Narang A, Bhakoo ON. Predictive perinatal score in the diagnosis of neonatal sepsis. J Trop Pediatr. 1994;40(6):365-368.
Crossref - Zea-Vera A, Ochoa TJ. Challenges in the diagnosis and management of neonatal sepsis. J Trop Pediatr. 2015;61(1):1-13.
Crossref - Shrestha RK, Rai SK, Khanal LK, Manda PK. Bacteriological study of neonatal sepsis and antibiotic susceptibility pattern of isolates in Kathmandu, Nepal. Nepal Med Coll J. 2013;15(1):71-73. PMID: 24592800
- Shah BA, Padbury JF. Neonatal sepsis. An old problem with new insights. Virulence. 2014;5(1):170-178.
Crossref - Okomo U, Akpalu ENK, Le Doare K, et al. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: a systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect Dis. 2019;19(11):P1219-P1234.
Crossref - Haque KN. Definitions of bloodstream infection in the newborn. Pediatr Crit Care Med. 2005;6(3):S45-S49.
Crossref - Isenberg H. Clinical Microbiology Procedure Handbook. 2nd Edition. ASM Press, Washington DC.; 2004.
- IBM Corp. Released. IBM SPSS Statistics for Mac, Version 21.0. 2012. 2012. https://www.scirp.org/(S(vtj3fa45qm1ean45vvffcz55))/reference/ReferencesPapers.aspx?ReferenceID=1311807
- Jain NK, Jain VM, Maheshwari S. Clinical profile of neonatal sepsis. Kathmandu Univ Med J (KUMJ). 2003;1(2):117-120
- English M, Ngama M, Mwalekwa L, Peshu N. Signs of illness in Kenyan infants aged less than 60 days. Bull World Health Organ. 2004;82(5):323-329. PMID: 15298222
- The Young Infant Clinical Study Group. Clinical signs that predict severe illness in children under age 2 months: a multicentre study. Lancet. 2008;371(9607):P135-142.
Crossref - Nawshad Uddin Ahmed ASM, Azad Chowdhury MAK, Hoque M, Darmstadt GL. Clinical and bacteriological profile of neonatal septicemia in a tertiary level pediatric hospital in Bangladesh. Indian Pediatr. 2002;39(11):1034-1039. https://pubmed.ncbi.nlm.nih.gov/12466574/
- Mugalu J, Nakakeeto MK, Kiguli S, Kaddu-Mulindwa DH. Aetiology, risk factors and immediate outcome of bacteriologically confirmed neonatal septicaemia in Mulago hospital, Uganda. Afr Health Sci. 2006;6(2):120-126.
Crossref - Shitaye D. Neonatal Sepsis: Bacterial Etiologic etiologic Agents agents and Their their Antibiotic antibiotic Susceptibility susceptibility Pattern pattern in Tikur Anbessa University Hospital, Addis Ababa, Ethiopia. Addis Ababa Univ. 2008. (Thesis) http://213.55.95.56/handle/123456789/5116?show=full
- Meremikwu MM, Nwachukwu CE, Asuquo AE, Okebe JU, Utsalo SJ. Bacterial isolates from blood cultures of children with suspected septicaemia in Calabar, Nigeria. BMC Infect Dis. 2005;5:110.
Crossref - Shrestha P, Das B, Bhatta N, et al. Clinical and Bacteriological Profiles of Blood Culture Positive Sepsis in Newborns. J Nepal Paediatr Soc. 2007;27(2):64-67.
Crossref - Nagaveni P, Suchitra D. Clinical profile of neonates admitted with sepsis – a tertiary care experience. IOSR J Dent Med Sci. 2016;15(4):49-56.
Crossref - Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza- Tanzania. BMC Pediatr. 2010;10:39.
Crossref - Varsha, Rusia U, Sikka M, Faridi MMA, Madan N. Validity of hematologic parameters in identification of early and late-onset neonatal infection. Indian J Pathol Microbiol. 2003;46(4):565-568. PMID: 15025344
- Wu JH, Chen CY, Tsao PN, Hsieh WS, Chou HC. Neonatal Sepsis: A 6-Year Analysis in a Neonatal Care Unit in Taiwan. Pediatr Neonatol. 2009;50(3):88-95.
Crossref - Costello A, Manandhar D. Improving Newborn Infant Health in Developing Countries. 2000.
Crossref - Sah R, Khadka S, Shrestha GS, et al. Detection of Pan drug resistance OXA-48 producing Providencia in an ICU patient for the first time in Nepal. Antimicrob Resist Infect Control. 2019;8:155.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.