ISSN: 0973-7510
E-ISSN: 2581-690X
Fusarium pathogens and their mycotoxins are considered as the main threats to cereal production and food safety worldwide. However, due to the constant discovery of new Fusarium species especially along with mycotoxin production profile differentiation in certain species, efforts on their species composition, geographical distribution, and chemotype proportion are urgently required. In the Fusarium goolgardi species, two distinct trichothecene (TB) genotype populations have recently been identified. Previous studies have shown that the structural variance of TBs biosynthesized by the two genotypes is attributed to the Tri1 gene. Polymorphisms of Tri1 gene from type A TB-producers were investigated in different Fusarium species in the current study. According to these DNA sequence variations identified in Tri1 gene sequences, a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) based diagnostic approach for the differentiation of T-2 and 4,15-diacetoxyscirpenol (DAS) genotypes in F. goolgardi was successfully developed. The PCR-RFLP assay will facilitate the studies on geographic distribution, frequency and other aspects of the two genotypes within F. goolgardi species.
Type A Trichothecenes, Genotype Identification, Fusarium mycotoxins, Fusarium goolgardi
Fusarium is one of the most economically destructive fungal genera with many species that cause variety of plant diseases. Members of Fusarium are the primary cause of fusarium head blight (FHB) or scab, a catastrophic disease that affects cereal production worldwide.1-6 FHB epidemics in North America and China have resulted in huge economic losses.1,4-9 As an example in the United States, FHB has caused more than 3 billion dollars losses in crop (include wheat and barley) since 1990s.1,6 Likewise, frequent epidemics and huge yield losses induced by FHB were also reported in China, with the Yangtze River basin to be most severe.4,5,7-10
The fungi from Fusarium genera are also of concern because they can produce variety of mycotoxins such as TBs, a series of non-volatile sesquiterpenes have a common core skeleton. Currently, over two hundred TBs have been found, posing a serious risk to the safety of food and feed.11 Among these Fusarium secondary metabolites, type A and type B TBs are the main contaminants of small grains and the two group compounds have aroused considerable public concern worldwide.12
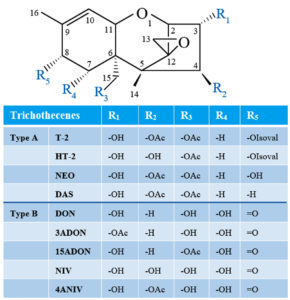
Type A and type B TBs are mainly characterized by the presence or absence of a ketone functional group at carbon 8 (abbreviated as C-8). In Type A TBs, there is an ester functional group, or a hydroxyl, or no substituent at all at C-8 position of the core TB molecule, and T-2, neosolaniol (NEO), and 4,15-diacetoxyscirpenol (DAS), respectively, are representative of this group (Figure 1).13,14 Whereas type B TBs are distinguished from type A by the appearance of a ketone functional group at C-8, and they are represented by deoxynivalenol (DON), nivalenol (NIV), as well as their acetylated derivatives (Figure 1).14-16 Fusarium TBs of particular concern are type A groups, which are very poisonous.17 It has been demonstrated that Type A TBs are strong inhibitors of DNA, RNA, and protein synthesis; they can cause immunosuppressive and pathological changes in liver cells.18,19 Furthermore, these toxins can induce DNA fragmentation characteristic of apoptosis and even death.20,21
Figure 1. Structures of common Type A and B trichothecenes (NEO, neosolaniol; DAS, 4,15-diacetoxyscirpenol; DON, deoxynivalenol; 3ADON, 3-acetyl-deoxynivalenol; 15ADON, 15-acetyl-deoxynivalenol; NIV, nivalenol; 4ANIV, 4-acetyl-nivalenol; OAc, acetyl function; OIsoval, isovalerate)
Traditionally, chemotyping of Fusarium strains have been carried out using gas chromatography/mass spectroscopy which are relatively time-consuming and expensive.22 Nevertheless, PCR based molecular genetic assay approaches (genotype assays) facilitate the quick screening of Fusarium strain toxin potential. Different genotype analysis methods for the identification of type B TB producing Fusarium species have been developed during the last two decades.23-28 All these methods are based on the polymorphisms of the specific Tri genes which are involved in TB biosynthesis. Genotyping is a useful tool for predicting TB production potential of Fusarium strains, and is commonly used for Fusarium graminearum species complex (FGSC)strain genotyping studies.22,29,30 High-throughput diagnostic approaches for type A TB producers, on the other hand, have yet to be established.31
Fusarium goolgardi is a new species recently identified and the members of this species can produce type A TBs.32,33 According to a recent investigation by Rocha LO et al., F. goolgardi populations have at least two TB genotypes: DAS-NEO-T2 genotype (abbreviated as T-2 genotype) and DAS genotype.33 Available data demonstrates that the enzyme encoded by Tri1 is required to catalyze the hydroxylation reaction of type A TBs at C-8 position, and thus it is involved in the biosynthesis of T-2, DAS, and NEO.17,33 Current study focused on: (1) the polymorphisms of Tri1 gene from type A TB producers; (2) development of molecular genotyping methods for the diagnosis of T-2 and DAS genotypes in F. goolgardi based on their Tri1 gene polymorphisms.
Tri1 gene sequence analysis
The Tri1 gene nucleotide sequences of different Fusarium species that produce type A TBs were obtained from GenBank database and evaluated for polymorphism. In total, eighteen Tri1 DNA sequences from four Fusarium species, including F. goolgardi (8 strains: RBG5411, 6914, 6915, 5417, 5419, 5420, 5421, and 5422), F. langsethiae (3 strains: NRRL53410, 53417, and 53439), F. sibiricum (2 strains: NRRL53421 and 53427), and F. sporotrichioides (5 strains: NRRL3299, 53434, 26924, 29977, and 29978), were retrieved from the NCBI GenBank and submitted to multiple sequence alignment analysis with Clustal W (version 2.0).34 The species were chosen for their ability to synthesize type A TBs, and all of the strains included in the study had previously been determined by chemical analyses.33,35,36 Table contains complete information on all representative sequences.
Fusarium strain culture and DNA extraction
Mycelium of Fusarium strains grown on sterile glass-membrane paper overlaying potato dextrose agar, was scraped from the surface cultures and ground to a fine powder in the presence of liquid nitrogen. Homogenized mycelia were suspended in 650 μL pre-heated (65°C) CTAB buffer, mix well and then incubate in a water bath (65°C, 40 min). After incubation, total genomic DNA was extracted as previously reported.28 Finally, DNA was diluted to ~50 ng/µL and kept frozen until required.
PCR amplification
Amplifications were performed in a final volume of 20 μL containing 1 × PCR buffer, 0.25 mmol/L of each dNTP, 0.2 mmol/L of primer A-Tri1F and A-Tri1R, and around 50 ng genomic DNA. The reaction was programmed as follows: 94°C 4 min; 30 cycles of 94°C 30 s, 60°C 20 s, 72°C 35 s; and 5 min final extension at 72°C. PCR products (2 μL) were separated through a 1.5% agarose gel electrophoresis and visualized under ultraviolet light after stained with nucleic acid fluorescent stain reagent (Transgen Biotech, Beijing, China). The PCR product sizes were evaluated according to known DNA standards.
Enzyme digestion
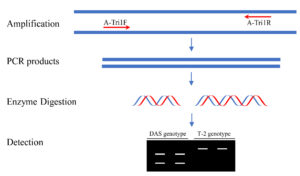
The Tri1 amplicon was subsequently digested with BfaI (NEB, Ipswich, MA, USA). The enzyme digestion was carried out in a 20 μL mixture containing 2 μL 10 × rCutSmart buffer, 3 μL Tri1 amplicon, 1 μL BfaI (10 U/μL), and 14 μL water. The digestion reaction was performed in a thermal cycler as previously described.31 Immediately after digestion, the products were detected by electrophoresis on 1.5% agarose gel. The approach is illustrated in Figure 2.
DNA polymorphism of Tri1 gene in Fusarium
In this study, we first compared the sequences of Tri1 homologs from three F. langsethiae (NRRL53410, 53417, 53439), two F. sibiricum (NRRL53421, 53427), and five F. sporotrichioides (NRRL3299, 53434, 26924, 29977, and 29978). The full-length Tri1 DNA sequences of these ten strains are varied from 1857 bp to 1860 bp with four introns, while the coding sequence is 1629 bp in all of them. Differences in DNA length of the intact Tri1 genes are due primarily to one and two nucleotides insertion/deletion in the third and fourth introns, respectively.
The coding sequences of these ten Tri1 DNA sequences were further subjected to alignment assays, and the results indicated that their identity ranged from 96.07% to 100%. The Tri1 coding sequences of F. sibiricum strain NRRL53421 is identical to NRRL53427, and identical coding sequences were observed in the three F. langsethiae strains NRRL53410, 53417, and 53439. A total of 109 single-nucleotide polymorphisms were found in the coding sequences of the examined 10 Tri1 sequences, while no other major differences were observed.
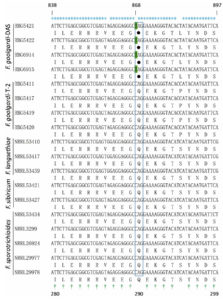
Portions of Tri1 sequence from eight F. goolgardi strains with different genotypes were included for subsequent polymorphism analysis and primer design. Nucleotide sequences of Tri1 gene DNA sequences from four F. goolgardi strains (RBG5411, 5417, 5419, and 5420) with T-2 genotype and four F. goolgardi strains (RBG6914, 6915, 5421, 5422) with DAS genotype were aligned against reference sequences from the above-mentioned F. langsethiae, F. sporotrichioides, and F. sibiricum strains (Table). As shown in Figure 3, Tri1 sequences in F. goolgardi strains with the DAS genotype differed significantly from those in T-2 producers. There is a nonsense mutation occurred at position 868 (C-to-T transition) in the middle of the Tri1 coding region, resulting in a premature stop codon and the failure of normal transcription and translation of Tri1 gene; therefore, it is considered as a pseudo-gene. While the Tri1 sequences from T-2-producing F. goolgardi strains did not exhibit the nonsense mutations, which generally will expressed normally.
Table:
Species, strains, genotypes, and sequence accession numbers of Fusarium Tri1 genes
Species |
Strains |
Geno-types |
Accession Numbers |
References |
|---|---|---|---|---|
F. goolgardi |
RBG5411 |
T-2 |
KT597908 |
Rocha LO et al. |
F. goolgardi |
RBG5417 |
T-2 |
KT597907 |
Rocha LO et al. |
F. goolgardi |
RBG5419 |
T-2 |
KT597906 |
Rocha LO et al. |
F. goolgardi |
RBG5420 |
T-2 |
KT597905 |
Rocha LO et al. |
F. goolgardi |
RBG5421 |
DAS |
KT597915 |
Rocha LO et al. |
F. goolgardi |
RBG5422 |
DAS |
KT597916 |
Rocha LO et al. |
F. goolgardi |
RBG6914 |
DAS |
KT597913 |
Rocha LO et al. |
F. goolgardi |
RBG6915 |
DAS |
KT597914 |
Rocha LO et al. |
F. langsethiae |
NRRL53410 |
T-2 |
HQ594538 |
Yli-Mattila et al. |
F. langsethiae |
NRRL53417 |
T-2 |
HQ594539 |
Yli-Mattila et al. |
F. langsethiae |
NRRL53439 |
T-2 |
HQ594543 |
Yli-Mattila et al. |
F. sibiricum |
NRRL53421 |
T-2 |
HQ594540 |
Yli-Mattila et al. |
F. sibiricum |
NRRL53427 |
T-2 |
HQ594541 |
Yli-Mattila et al. |
F. sporotrichioides |
NRRL26924 |
T-2 |
HQ594535 |
Yli-Mattila et al. |
F. sporotrichioides |
NRRL29977 |
T-2 |
HQ594536 |
Yli-Mattila et al. |
F. sporotrichioides |
NRRL29978 |
T-2 |
HQ594537 |
Yli-Mattila et al. |
F. sporotrichioides |
NRRL53434 |
T-2 |
HQ594542 |
Yli-Mattila et al. |
F. sporotrichioides |
NRRL3299 |
T-2 |
AY040587 |
Meek et al. |
Figure 3. Alignment analysis of the nucleotides and corresponding protein sequences of partial Tri1. The conserved nucleotides and amino acids are indicated above and under the panel by * and †, respectively. The nonsense mutation sites in the CDS position 868 (C to T) of the F. goolgardi DAS strains indicated with green background, and the corresponding stop codon is represented by a solid circle (referred to strain NRRL29978)
Primer design
The alignment allowed designing a pair of primers, A-Tri1F (5′-GTAGCAAATCACCTACGCAGAT-3′) and A-Tri1R (5′-GAGGACGCATTCTCGTATATCT-3′). The expected amplicon size is 628 bp for all the type A TB-producing Fusarium strains, including F. sporotrichioides, F. goolgardi, F. sibiricum, and F. langsethiae.
PCR-RFLP approach for genotyping of F. goolgardi
A PCR-RFLP protocol targeting on the Tri1 gene variations was designed to distinguish between DAS and T-2 genotypes (Figure 2). Due to a single-nucleotide mutation, the fragment amplified with A-Tri1F/R from the F. goolgardi DAS strains have been digested into two different fragments of varying sizes by BfaI, MaeI, MthZI, and RmaI (the four restriction endonucleases recognize the same specific nucleotide sequence “CTAG”). However, the amplicons from all the other strains, including the F. goolgardi T-2 strains, can’t be digested by any of these restriction endonucleases.
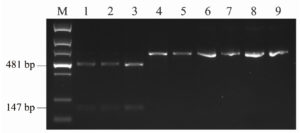
To evaluate the reliability of the PCR-RFLP approach for identifying DAS and T-2 genotype strains, strains from F. goolgardi, F. langsethiae, F. sibiricum, and F. sporotrichioides were tested. TBs produced by these strains have been reported previously.33,35,36 As shown in Figure 4, after digestion with BfaI, the amplicons from F. goolgardi DAS strains were cut into two fragments with different sizes, 147 bp and 481 bp, respectively, in length. On the other hand, a single 628 bp band was observed from the amplicons of T-2 producers. The unique DNA profiles obtained from DAS and T-2 genotype strains generated by PCR-RFLP assays suggest that this method can be used for differentiation of these two type A TB producers in F. goolgardi populations. Furthermore, according to our analyses, the molecular genotyping detection method developed in the current study may also can be extended to identify DAS and T-2 genotypes in other species, such as F. langsethiae, F. sibiricum, and F. sporotrichioides.
Figure 4. The restriction enzyme digestion products of Fusarium strains amplified by primer set A-Tri1F/R. M, DNA ladder marker; Lane 1 to lane 3, F. goolgardi strains with DAS genotype: RBG6914, 6915, and 5421; Lane 4 to lane 5, F. goolgardi strains with T-2 genotype: RBG5411 and 5419; Lane 6, F. langsethiae strain NRRL53410 (T-2 producer); Lane 7, F. sibiricum strain NRRL53421 (T-2 producer); Lanes 8 and 9, F. sporotrichioides strains NRRL29978 and NRRL3299 (T-2 producers), respectively
TBs’ structure diversity is caused by the variability of TB biosynthesis gene functions. As a result, Tri genes are often utilized as markers to assess the probable ability of Fusarium strains to produce TBs using PCR-based assays.22-29 As we know, Tri1 and Tri16 genes are critical for the hydroxylation reaction and its immediate acylation of the TB skeleton structure at C-8 position in the biosynthetic pathway of T-2, respectively. However, at C-8 of the DAS, there are just two hydrogen atoms, and no further modification occurred. Thus, compared with T-2 producers, there is probably a nonfunctional Tri1 gene or at least the gene can’t be expressed normally in Fusarium strains which can only produce DAS toxin.
The occurrence of nonsense mutation in the coding region of Tri1 gene from F. goolgardi strains with a DAS genotype renders the gene non-functional, thus leading to the production of TB homologues without C-8 modifications.17,33 The findings also show that other polymorphisms may have occurred in the DAS lineage, possibly resulting in Tri1 gene dysfunction. The precise scenario that resulted in these mutations in the Tri1 gene is difficult to pinpoint; nonetheless, such changes can be exploited to distinguish between DAS and T-2 producers in the Fusarium genera.
Only one genotype has been documented for F. langsethiae, F. sporotrichioides, and F. sibiricum, and strains from the three species can produce T-2, as well as other type A TBs, including DAS and NEO.18,20,33,35-37 However, the discovery of two different genotypes, DAS and T-2 types, within F. goolgardi strains has just been reported. The results from Rocha LO et al. demonstrated that some strains of F. goolgardi produced DAS and NEO in addition to T-2, while other strains produced only DAS.33
In the current study, a PCR-based approach was developed for the diagnosis of DAS and T-2 genotypes in F. goolgardi. In contrast to T-2 genotypes, a nonsense mutation occurred at position 868 (C-to-T transition) in the Tri1 gene of the DAS genotype F. goolgardi strains. This mutation resulted in the emergence of a new enzyme cleavage site, C*TAG (* denotes the cleavage site), which can be recognized by restriction endonucleases BfaI, MaeI, MthZI, and RmaI. Although a small number of Fusarium strains were evaluated with our PCR-RFLP method, findings reported in Rocha LO et al. provide significant support to our results.33
Different genotyping methods are extensively applied in species identification, molecular marker screening, and genetic diversity analysis. These fingerprinting techniques consider information distributed over the whole genome of an organism and generally permit discrimination of Fusarium strains with different phenotypes like different species or different TB profiles. Among these techniques, RFLP is famous for its robustness, reproducibility, and reliability.38 Recently, four sets PCR-RFLP diagnostic methods for the differentiation of type B TB- and NX-producing FGSC were successfully developed by us relying on polymorphisms identified in Tri1 sequences.31 As far as we know, though, this is the first report on the genotyping analysis of T-2 and DAS genotypes in F. goolgardi. Further investigations are needed to determine the fungi’s temporal and spatial distribution. The PCR-based RFLP assay developed in this work will make easier and faster the investigations on temporal and spatial distribution of DAS and T-2 populations.
Fusarium goolgardi is a recently identified Fusarium species.32 Therefore, further research will be necessary to track and monitor the two genotypes’ geographic distribution and spread as well as assess any potential variations in their competitive capacities, including pathogenicity and environmental adaptability. Moreover, the divergence of TB genotype within the F. goolgardi species also highlights the need to re-evaluate genotype and population diversity of the economically destructive fungal pathogens in Fusarium genera throughout different agricultural ecosystems worldwide.
On the other hand, subsequent identification of mutations (premature stop codons and frameshift) in Tri16 coding region in F. goolgardi strains carrying the DAS genotype give rise to new questions and perspectives that whether DAS genotype strains contain a functional Tri16 gene exist or not in nature.33 If so, what kind of TBs produced by such strains? It is still unclear whether strains that primarily produce NEO or coexistences of DAS and NEO, however, lack T-2 toxin exist or not.17 Taking this into account, we recently conducted a comprehensive and systematic analysis of the advances on type A TB biosynthesis in Fusarium species, with respective to their biosynthetic pathway, gene evolution and other concepts, and the potential biosynthetic pathways for NEO and DAS toxins were proposed.17 Based on the previous findings and current questions, further research is needed to discover the biochemical alterations or genetic basis changes in TB biosynthesis and related molecular regulation mechanisms. The settlement of these issues will undoubtedly give greater impetus to the understanding of evolution of secondary metabolites biosynthesis in Fusarium species, and provide new insights on the control and prevention of mycotoxins in food safety.
Identification of Fusarium strain toxin potential is essential with respect to food safety issues due to TBs’ different toxicological effects. Previously, two distinct TB genotype groups, namely T-2 genotype and DAS genotype, were found in the species of F. goolgardi. In this study, the polymorphisms of Tri1 gene from the two kinds of TB producers were investigated. A PCR-RFLP assay method was successfully developed to distinguish T-2 and DAS producers within F. goolgardi. This promising diagnostic method can be used for high-throughput genotype analysis of F. goolgardi strains as a step forward for plant disease management, TBs assessment and control in agriculture. Besides, this molecular diagnostic technology will facilitate the studies on its epidemiology, host range, and geographical distribution of the novel species.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was funded by the National Natural Science Foundation of China [grant numbers 31871896 and 32060592] and Shanghai Science and Technology Innovation Action Plan [grant numbers 23N31900500 and 23ZR1455700].
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Shanghai Academy of Agricultural Sciences, Shanghai, China.
- Windels CE. Economic and social impacts of Fusarium head blight: changing farms and rural communities in the northern great plains. Phytopathology. 2000;90(1):17-21.
Crossref - Bai GH, Shaner G. Management and resistance in wheat and barley to Fusarium head blight. Annu Rev Phytopathol. 2004;42:135-161.
Crossref - Goswami RS, Kistler HC. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol. 2004;5(6):515-525.
Crossref - Zhang J, Li H, Dang F, et al. Determination of the trichothecene mycotoxin chemotypes and associated geographical distribution and phylogenetic species of the Fusarium graminearum clade from China. Mycol Res. 2007;111(Pt 8):967-975.
Crossref - Qu B, Li H, Zhang J, et al. Geographic distribution and genetic diversity of Fusarium graminearum and F. asiaticum on wheat spikes throughout China. Plant Pathol. 2008;57(1):15-24.
Crossref - McMullen M, Bergstrom GC, De Wolf E, et al. A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis. 2012;96(12):1712-1728.
Crossref - Wang J, Ndoye M, Zhang J, Li H, Liao Y. Population structure and genetic diversity of the Fusarium graminearum species complex. Toxins. 2011;3(8):1020-1037.
Crossref - Zhang H, Van der Lee T, Waalwijk C, et al. Population analysis of the Fusarium graminearum species complex from wheat in China show a shift to more aggressive isolates. PLoS One. 2012;7(2):e31722.
Crossref - Chen Y, Kistler HC, Ma Z. Fusarium graminearum trichothecene mycotoxins: biosynthesis, regulation, and management. Annu Rev Phytopathol. 2019;57:15-39.
Crossref - Zhang J, Wang J, Gong A, et al. Natural occurrence of Fusarium head blight, mycotoxins and mycotoxin-producing isolates of Fusarium in commercial fields of wheat in Hubei. Plant Pathol. 2013;62(1):92-102.
Crossref - Zingales V, Fernandez-Franzon M, Ruiz M. Occurrence, mitigation and in vitro cytotoxicity of nivalenol, a type B trichothecene mycotoxin-updates from the last decade (2010-2020). Food Chem Toxicol. 2021;152:112182.
Crossref - Qiu J, Xu J, Shi J. Fusarium toxins in Chinese wheat since the 1980s. Toxins. 2019;11(5):248.
Crossref - Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci Biotechnol Biochem. 2007;71(9):2105-2123.
Crossref - McCormick SP, Stanley AM, Stover NA, Alexander NJ. Trichothecenes: from simple to complex mycotoxins. Toxins. 2011;3(7):802-814.
Crossref - Varga E, Wiesenberger G, Hametner C, et al. New tricks of an old enemy: isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ Microbiol. 2015;17(8):2588-2600.
Crossref - Kelly AC, Proctor RH, Belzile F, et al. The geographic distribution and complex evolutionary history of the NX-2 trichothecene chemotype from Fusarium graminearum. Fungal Genet Biol. 2016;95:39-48.
Crossref - Wang J, Zhang M, Yang J, Yang X, Zhang J, Zhao Z. Type A trichothecene metabolic profile differentiation, mechanisms, biosynthetic pathways, and evolution in Fusarium species-a mini review. Toxins. 2023;15(7):446.
Crossref - Torp M, Nirenberg HI. Fusarium langsethiae sp. nov. on cereals in Europe. Int J Food Microbiol. 2004;95(3):247-256.
Crossref - Wu J, Jing L, Yuan H, Peng S. T-2 toxin induces apoptosis in ovarian granulosa cells of rats through reactive oxygen species-mediated mitochondrial pathway. Toxicol Lett. 2011;202(3):168-177.
Crossref - Torp M, Langseth W. Production of T-2 toxin by a Fusarium resembling Fusarium poae. Mycopathologia. 1999;147(2):89-96.
Crossref - Rocha O, Ansari K, Doohan FM. Effects of trichothecene mycotoxins on eukaryotic cells: a review. Food Addit Contam. 2015;22(4):369-378.
Crossref - Wang J, Zhao Z, Yang X, et al. In Sabuncuoglu S (eds.), Mycotoxins and Food Safety, 1th Ed. Chapter: Fusarium graminearum species complex and trichothecene genotype. IntechOpen, London, United Kingdom. 2019.
Crossref - Lee T, Oh DW, Kim HS, et al. Identification of deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae by using PCR. Appl Environ Microbiol. 2001;67(7):2966-2972.
Crossref - Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O’Donnell K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc Natl Acad Sci USA. 2022;99(14):9278-9283.
Crossref - Chandler EA, Simpson DR, Thomsett MA, Nicholson P. Development of PCR assays to Tri7 and Tri13 trichothecene biosynthetic genes, and characterisation of chemotypes of Fusarium graminearum, Fusarium culmorum and Fusarium cerealis. Physiol Mol Plant Pathol. 2003;62(6):355-367.
Crossref - Jennings P, Coates ME, Walsh K, Turner JA, Nicholson P. Determination of deoxynivalenol- and nivalenol-producing chemotypes of Fusarium graminearum isolated from wheat crops in England and Wales. Plant Pathol. 2004;53(5):643-652.
Crossref - Li HP, Wu AB, Zhao CS, Scholten O, Loffler H, Liao YC. Development of a generic PCR detection of deoxynivalenol- and nivalenol-chemotypes of Fusarium graminearum. FEMS Microbiol Lett. 2005;243(2):505-511.
Crossref - Wang J, Li H, Qu B, et al. Development of a generic PCR detection of 3-acetyldeoxynivalenol-, 15-acetyldeoxynivalenol- and nivalenol-chemotypes of Fusarium graminearum clade. Int J Mol Sci. 2008;9(12):2495-2504.
Crossref - Pasquali M, Giraud F, Brochot C, Cocco E, Hoffman L, Bohn T. Genetic Fusarium chemotyping as a useful tool for predicting nivalenol contamination in winter wheat. Int J Food Microbiol. 2010;137(2-3):246-253.
Crossref - Covarelli L, Beccari G, Prodi A, et al. Fusarium species, chemotypes characterization and trichothecene contamination of durum and soft wheat in an area of central Italy. J Sci Food Agr. 2015;95(3):540-551.
Crossref - Gao M, Zhang M, Zhang J, Yang X, Abdallah MF, Wang J. Phylogenetic variation of Tri1 gene and development of PCR-RFLP analysis for the identification of NX genotypes in Fusarium graminearum species complex. Toxins. 2023;15(12):692.
Crossref - Laurence MH, Walsh JL, Shuttleworth LA, et al. Six novel species of Fusarium from natural ecosystems in Australia. Fungal Diversity. 2016;77(1):349-366.
Crossref - Rocha LO, Laurence MH, Proctor RH, McCormick SP, Summerell BA, Liew ECY. Variation in type A trichothecene production and trichothecene biosynthetic genes in Fusarium goolgardi from natural ecosystems of Australia. Toxins. 2015;7(11):4577-4594.
Crossref - Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947-2948.
Crossref - Meek IB, Peplow AW, Ake CJr, Phillips TD, Beremand MN. Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. Appl Environ Microbiol. 2003;69(3):1607-1613.
Crossref - Yli-Mattila T, Ward TJ, O’Donnell K, et al. Fusarium sibiricum sp. nov, a novel type A trichothecene-producing Fusarium from northern Asia closely related to F. sporotrichioides and F. langsethiae. Int J Food Microbiol. 2011;147(1):58-68.
Crossref - Thrane U, Adler A, Clasen P, et al. Diversity in metabolite production by Fusarium langsethiae, Fusarium poae, and Fusarium sporotrichioides. Int J Food Microbiol. 2004;95(3):257-266.
Crossref - Baffoni L, Stenico V, Strahsburger E, et al. Identification of species belonging to the Bifidobacterium genus by PCR-RFLP analysis of a hsp60 gene fragment. BMC Microbiol. 2013;13:149.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.