ISSN: 0973-7510
E-ISSN: 2581-690X
Dermatophytosis is the superficial infection of keratinized tissue like skin, hair, and nails, in humans and animals, by a group of closely related fungi known as dermatophytes. Phenotypic identification of dermatophytes, especially through classical methods can be difficult and uncertain at times, especially when differentiating species with overlapping characteristics. Alternative identification methods based on amplification and sequence analysis of the highly polymorphic internal transcribed spacer (ITS) sequences flanking the 5.8S ribosomal RNA gene has proven to be quite sensitive and reliable. The objective of our study was to compare the phenotypic and the ITS sequencing-based methods for the identification of clinically isolated dermatophyte specimens from Puducherry, India. A total of 13 clinical samples from 39 suspected cases were found positive for dermatophytes using KOH/DMSO preparations. Specimens were subsequently cultured in Sabouraud dextrose agar (SDA) supplemented with chloramphenicol, gentamicin, and cycloheximide. Dermatophytes were identified based on culture characteristics and microscopic examination in lactophenol cotton blue preparations. ITS sequencing was additionally performed after PCR amplification for species identification. Identification based on phenotype through microscopy and culture methods confirmed infections with Trichophyton mentagrophytes (n = 11), T. rubrum (n = 1), and Microsporum gypseum (n = 1). The strains were confirmed by ITS sequencing without any discrepancy with phenotypic identification. Identification of common dermatophytes based on phenotypic characteristics may be used as a reliable method of diagnosis where sophisticated methods like ITS sequencing and PCR are unavailable.
Dermatophytosis, ITS sequencing, Trichophyton mentagrophytes, Trichophyton rubrum, Microsporum gypseum, dermatophytes
Dermatophytes are fungi that cause superficial, painless but often pruritic infections of the skin, hair, and nails known as dermatophytosis or more commonly as ringworm or tinea infections1. They can infect humans and animals alike. Infections are generally limited to the non-living stratum corneum of the epidermis.2,3 Occasionally, however, these fungi can become invasive, causing a deeper and disseminated infection, which can become a cause of concern.4
Widespread lesions may negatively impact the quality of life socially, occupationally, and psychologically.5
Infection is established as the fungus colonizes on the outer surface of keratinized tissues like the skin, nails, and hair. The arthroconidia attach to the epidermis using specific proteins expressed on the fungal surface and release proteases like subtilisin 3.6-9 Within a few hours, germination and hyphal growth commence.10 Between 24 hours to 3 days, the hyphae invade the stratum corneum and start degrading the corneodesmosomes, lipid matrix, and keratin.11 As the hyphae penetrate deeper into the stratum corneum, disruption of tight junctions leads to the loss of epidermal barrier integrity, release of proinflammatory cytokines and antimicrobial peptides by keratinocytes, and immune recruitment. In a majority of clinical cases, pruritic lesions are observed. Scratching of the lesions often results in secondary bacterial infections like with Staphylococcus aureus. Successful control of dermatophytosis alleviates the itch and reduces secondary infections associated with scratching behavior.12
Dermatophytosis is fairly common but neglected among the Indian population. Current reports from India estimate a prevalence of 36.6-78.4%.13,14 There has been an unusual increase in dermatophytosis cases presenting with larger than normal lesions, ring within ring lesions, multiple site lesions, and corticosteroid modified lesions. This unexpected change in the clinical presentation of dermatophytosis, compounded by the difficulty in diagnosis, has become a matter of concern among dermatologists.15 Despite the increase in prevalence not only in India but also throughout the world, research in this area has been neglected.16 The current situation is exacerbated by more humid and warm climates, irresponsible use of topical corticosteroids that are easily available over the counter, alarming rise in the use of broad-spectrum antibiotics, increase in immunocompromised population, rampant use of antifungals in agro-industries, and antifungal drug resistance, especially terbinafine.17-19
Dermatophytosis is caused by asexual or imperfect fungi of three genera, namely, Trichophyton, Microsporum, and Epidermophyton. These anamorphic genera have about 40 species. Some zoophilic and geophilic species of Trichophyton and Microsporum can reproduce sexually and are classified as teleomorphs of the genus Arthroderma. Dermatophytosis, also known as tinea, is named after the anatomical location of the infection. Multiple anatomic locations may be infected by the same dermatophyte species and many different species may lead to clinically identical tinea lesions.1 Trichophyton rubrum has a global occurrence while others may vary in their geographic distribution. However, reports from different parts of India indicate that the distribution of dermatophytes in India does not vary much geographically.14 Identifying the etiologic agent to the species level can provide insight into the epidemiological spread of the disease and allow the adoption of specific treatment regimens for better management of infections. It is also imperative to identify dermatophyte-like non-pathogenic fungi that may also grow from dermatophytic lesions. Additionally, dermatophytes must be differentiated from non- dermatophytic fungi that cause dermatophyte-like infections known as onychomycosis caused by Neoscytalidium dimidiatum.
Dermatophytes can be identified to the genus and sometimes to the species level based on phenotypic differences. A quick confirmatory test is the direct microscopic observation of scrapings from a suspected infection when mixed with KOH and DMSO.20 The presence of fungal hyphae is indicative of dermatophytosis. Sabouraud dextrose agar (SDA) originally formulated by Sabouraud and subsequently modified by Emmons21 when used with chloramphenicol and cycloheximide can act as a selective media which inhibits the growth of other pathogenic fungi while promoting the growth of dermatophytes. The growth characteristics in SDA, dermatophytes test medium (DTM), and enriched dermatophytes medium (EDM) followed by urease utilization, hair perforation test, and microconidia and macroconidia morphology in lactophenol cotton blue (LPCB) mount can be used in identifying different dermatophytes. As reliable as these methods are, it has however been observed that the identification of dermatophytes can sometimes be difficult and uncertain due to variations in the isolates and overlapping characteristics between species.22 Molecular biology-based approaches like RFLP, PCR, and NGS have been used to differentiate closely related species. These techniques exploit the polymorphism in the internal transcribed spacers (ITS) flanking the gene encoding the 5.8S ribosomal RNA and have proven to be very sensitive and reliable in differentiating dermatophytes.23
This study compares the phenotypic and molecular identification based on ITS1 and ITS4 sequence analysis of clinical samples of dermatophytes collected in a tertiary care hospital in Puducherry, India. Further assessed was the relationship of clinical manifestations with gender and age and the possible correlation of clinical manifestations with the dermatophyte isolates.
The study was approved by the MGMC & RI Institutional Human Ethics Committee (project number: FACULTY_PROJECT/04/2019/06 dated 22/04/2019) as per Indian Council of Medical Research guidelines 2017 and was performed according to the WMA Declaration of Helsinki ethical principles for research on humans.
Participants
39 patients, aged between 13 and 69 years, clinically suspected of dermatophytosis at the dermatology outpatient department of Mahatma Gandhi Medical College and Research Institute (MGMC & RI), Puducherry, India, between May and October 2019, were recruited for the study. All patients with clinically diagnosed dermatophytosis, irrespective of their age and sex, who were not undergoing treatment for the same, were selected for the study. Patients already undergoing treatment for dermatophytosis were excluded.
Sampling Procedure
Patients clinically suspected of dermatophytosis were chosen for the study after informed consent. The infected area on the skin was cleaned with 70% ethanol and ensured complete dryness. Using a sterile dull scalpel, scrapings from the advancing margin of the lesion were collected in a sterile container. Any hair present in the scrapings was removed aseptically.
Microscopy and Culture
Fungal microscopic characteristics were observed in KOH-DMSO preparations.24 Scrapings were placed on a slide and a few drops of a solution of 20% KOH in 40% DMSO were added. A coverslip was carefully placed to cover the entire scrapings. All specimens were immediately examined by light microscopy, under low and high power, for unstained refractile fungal elements. Sabouraud dextrose agar (SDA) (HiMedia Laboratories, Mumbai, India) was used as a non-selective medium for the isolation of fungal pathogens.1 Plates with SDA supplemented with chloramphenicol and plates with SDA supplemented with chloramphenicol, gentamicin, and cycloheximide were simultaneously streaked with scraped specimens from patients. Each plate was inoculated in duplicates and incubated at 25 and 37°C. Positive cultures were examined daily for six weeks. The dermatophytes were identified based on their colony morphology and microscopic characteristics in the lactophenol cotton blue (LPCB) preparations using slide culture techniques.
DNA extraction
DNA was extracted from fungal cultures using DNAQuik™ reagent (Bioserve Biotechnologies (India) Pvt. Ltd) and quantified by the picogreen protocol. The quality of DNA was analyzed by electrophoresing 25-200 ng of DNA on a 1% agarose gel.
DNA amplification and ITS DNA Sequencing
The extracted DNA was used in a polymerase chain reaction (PCR) to amplify the internal transcribed spacer (ITS) region using ITS1 and ITS4 primers, as described by Gardes and Bruns. The PCR program included the following steps: initial denaturation at 94°C for 85 seconds, followed by 35 cycles of denaturation, annealing, and extension. For the initial first 13 cycles, the temperature and times for denaturation, annealing, and extension used were 95°C for 35 seconds, 55°C for 55 seconds, and 72°C for 45 seconds respectively and for cycles 14-26 and 27-35, the time for extension steps alone were lengthened to 120 and 180 seconds, within the two set of cycles respectively. After the completion of the thermocycling, the samples were incubated for 10 minutes at 72°C.25 The 400-900 bp amplicon was gel eluted and the product was sequenced by Sanger’s method. The sequence data were assembled and using the nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), sequences that produced significant alignments (percent identity ≥ 95%) with the query sequence were used in species identification.
Among 39 patients with suspected dermatophytosis, 13 were confirmed in culture; out of which, seven were females (53.84%) and six were males (46.15%) (Table 1). Microscopic examination in LPCB preparations was performed based on hyphae septation, conidiophore branching, microconidia clustering, and macroconidia shape. Microscopic and colony characterization revealed specimens of Trichophyton mentagrophytes, Trichophyton rubrum, and Microsporum gypseum.
Table (1):
Etiology of dermatophytosis.
| Etiological agent | Isolates | Male | patients | Female patients | ||
|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | |
| T. mentagrophytes | 11 | 84.61 | 5 | 38.46 | 6 | 46.15 |
| T. rubrum | 1 | 7.69 | 1 | 7.69 | 0 | 0 |
| M. gypseum | 1 | 7.69 | 0 | 0 | 1 | 7.69 |
| Total | 13 | 100 | 6 | 46.15 | 7 | 53.84 |
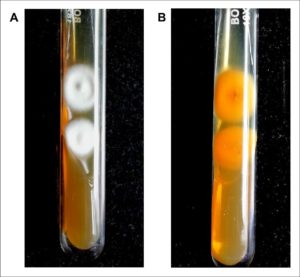
Fig. 1. Colony morphology of dermatophyte isolated from suspected clinical samples in SDA.
(A) Culture after 6-10 days shows fluffy white colonies with raised centers. (B) Reverse pigmentation was found to be yellowish-brown with a central reddish-brown patch. The morphology is consistent with Trichophyton mentagrophytes and was confirmed by DNA sequencing of the ITS regions (GenBank accession number MW454380).
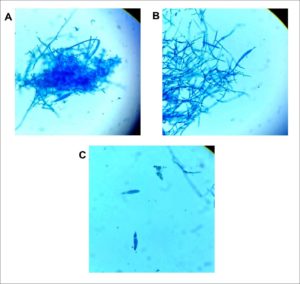
Fig. 2. Microscopic examination of LPCB stained dermatophyte identified as Trichophyton mentagrophytes. Magnification ×400. (A) Spherical conidia in clusters seen extending from septate hyphae and a macroconidium can be seen near the lower left center of the photo. (B) Sessile microconidia are seen along septate hyphae. (C) Two solitary cigar-shaped macroconidia showing three internal cells with thin walls and a smooth exterior. Identification was subsequently confirmed by DNA sequencing of the ITS regions (GenBank accession number MW454380).
Among the 13 patients, 11 presented with tinea corporis, one with tinea faciei, and one with tinea incognito infections. Trichophyton spp. was isolated from 12 patients (92.3%) (Fig. 1 and 2), while Microsporum gypseum was isolated from one patient (7.6%). Of the 12 Trichophyton positive cases, T. mentagrophytes was the predominant species isolated (91.66%), followed by T. rubrum (8.33%). Of the 11 patients with T. mentagrophytes isolates, 9 presented symptoms of tinea corporis (81.8%), one of tinea faciei (9.09%), and one of tinea incognito (9.09%). Both the patients with T. rubrum and Microsporum gypseum isolates presented with clinical symptoms of tinea corporis (Table 2). The ITS region of the DNA isolated from all the 13 culture isolates were amplified successfully by PCR. DNA sequencing of the ITS regions identified 11 cases as T. mentagrophytes (GenBank accession numbers: MW454380, MW454772, MW454799, MW454828, MW454864, MW463449, MW463454, MW463455, MW463457, MW463460, MW463459), one case as T. rubrum (GenBank accession number: MW454827), and one case as Nannizzia gypsea (previously known as Microsporum gypseum) (GenBank accession number MW454821) (Table 3). No discrepancies between the phenotypic and molecular identifications were observed.
Table (2):
Clinical manifestation of dermatophytosis.
| Clinical manifestation | Isolates | T. mentagrophytes | T. rubrum | M. gypseum | ||||
|---|---|---|---|---|---|---|---|---|
| No. of patients | % | No. of isolates | % | No. of isolates | % | No. of isolates | % | |
| Tinea corporis | 11 | 84.61 | 9 | 69.23 | 1 | 7.69 | 1 | 7.69 |
| Tinea faciei | 1 | 7.69 | 1 | 7.69 | 0 | 0 | 0 | 0 |
| Tinea incognito | 1 | 7.69 | 1 | 7.69 | 0 | 0 | 0 | 0 |
| Total | 13 | 100 | 11 | 84.61 | 1 | 7.69 | 1 | 7.69 |
Table (3):
Details of dermatophyte strains isolated with a correlation between phenotypic and molecular identification by DNA sequencing of the ITS region.
| No. | Age | Sex | Clinical diagnosis | Identification | ||
|---|---|---|---|---|---|---|
| Phenotypic | Molecular (ITS DNA sequencing) | GenBank accession numbers | ||||
| 1 | 16 | M | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW463460 |
| 2 | 20 | M | Tinea faciei | T. mentagrophytes | T. mentagrophytes | MW463459 |
| 3 | 22 | F | Tinea incognito | T. mentagrophytes | T. mentagrophytes | MW463457 |
| 4 | 32 | F | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW463455 |
| 5 | 33 | F | Tinea corporis | M. gypseum | Nannizzia gypsea | MW463454 |
| 6 | 37 | M | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW463449 |
| 7 | 37 | M | Tinea corporis | T. rubrum | T. rubrum | MW454864 |
| 8 | 37 | F | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW454828 |
| 9 | 39 | F | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW454827 |
| 10 | 43 | F | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW454821 |
| 11 | 48 | M | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW454799 |
| 12 | 60 | F | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW454772 |
| 13 | 69 | M | Tinea corporis | T. mentagrophytes | T. mentagrophytes | MW454380 |
Epidemiological studies on dermatophytosis have been carried out by different groups in India and indicate a countrywide prevalence.14 While most studies have indicated the primary etiologic agents to be T. mentagrophytes and T. rubrum, the geographical origin of the strains has remained difficult to ascertain. Increased travel and migration of people has been regarded as a contributing factor. Various tests based on histology, colony characteristics, ability to hydrolyze urea, hair perforation, PCR, RFLP, MALDI-TOF MS, and sequencing have been improved upon to detect different dermatophytes.22,26 While molecular techniques are often more reliable than conventional methods of identification, PCR methods have been shown to vary between investigators. This could be due to differences in the DNA extraction protocols, PCR primer design, and/or analysis of results.22 The polymorphism in the ITS1 and ITS2 regions flanking the 5.8S rDNA sequence has been used as a reliable marker for accurately differentiating dermatophyte species.27,28 Sequencing of ITS regions has been successfully used in the identification of closely related dermatophytes.29,30 A 2019 study from Senegal, Africa, used ITS sequencing to compare identification with morphological characteristics.31 Their results, however, indicated discrepancies in five out of 32 identifications. A 2015 study by an Iranian group had also reported discrepancies in their phenotypic versus genotypic results.32 The PCR- RFLP-based method, which has been used for molecular identification of dermatophytes, has also shown discrepancies in their observations.33,34 This could be because many dermatophytes exhibit overlapping morphological characteristics which makes phenotypic identification error-prone.
In the present study, we compared the phenotypic identification with the molecular identification of dermatophytes isolated from patients suffering from tinea corporis, tinea faciei, and tinea incognito. Samples collected from suspected patients were characterized microscopically as well as in SDA cultures. Phenotypic characterization was based on lactophenol cotton blue-stained morphology and colony characteristics. Molecular identification relied on the sequencing of ITS regions of ribosomal DNA. DNA was extracted from the fungal strains, and a specific ITS region on rDNA was amplified by PCR using primers designed for the flanking ITS1 and ITS4 regions. The 400-900 bp amplicons were sequenced and compared using NCBI BLAST to identify the fungal strain. Of the suspected 39 cases of dermatophytosis, 13 were confirmed by ITS sequencing after phenotypic identification. The predominant fungus isolated was Trichophyton and indicates a high prevalence of this fungus causing dermatophytosis in the rural population in and around Puducherry. This study agrees with earlier findings of dermatophytosis in India.13,35-40 However, unlike the previous studies, which have indicated a high prevalence of both T. mentagrophytes and T. rubrum (30-64%), our study reports an exceptionally high prevalence of T. mentagrophytes (84.61%) and low prevalence of T. rubrum (7.69%). A majority of patients (11/13) presented symptoms of tinea corporis, with the infection in males being similar to females (1:1.2). The most common age group to be infected by dermatophytes was 31-40 years (6/13). Interestingly, in this age group, the number of females with tinea corporis (4/6) was much higher than males (2/6). This higher incidence in females than earlier reported35 could be due to higher reporting as a result of the mitigation of social stigma in rural India. Other infections were of tinea faciei and tinea incognito. The etiological agent for both of which was found to be T. mentagrophytes. It is noteworthy that all our phenotypic identifications match the ITS sequence-based identifications. We have achieved a good correlation between phenotypic and molecular technique-based identification, as previously reported by Pryce et al.41 However, the comparatively smaller sample size (n = 13) may have precluded any possible erroneous identifications. This leaves scope for further investigations using larger sample sizes and more specific phenotypic tests like urease test, hair perforation test, and use of specific media like Lactritmel agar, Littman Oxgall agar, and Trichophyton agar, among others.
Superficial fungal infections are gaining significance in developing countries like India, and of the etiologic agents, dermatophytes are the most common. Due to their ability to metabolize keratin, they cause a range of pathological presentations. Though such infections are normally limited to the stratum corneum, they can become invasive and disseminate throughout the body, making it a matter of concern and not neglect. Dermatophyte skin infections may often resemble other non-fungal skin infections.42 and therefore correct knowledge of etiologic agents is critical to initiate timely treatment. Widespread dermatophytosis may affect one’s quality of social, psychological, and occupational life. Since no vaccine for dermatophytosis exists, spreading awareness about the disease, quick diagnosis, and timely treatment can help arrest the spread of the disease.
Our comparison of the phenotypic identification, based on KOH microscopy, SDA culture, and LPCB staining with the molecular identification, based on ITS amplicon sequencing, has revealed no discrepancy. Identification of common dermatophytes based on phenotypic characters may thus be regarded as a reliable method of diagnosis. However, phenotypic identification is often time-consuming and sometimes takes weeks to differentiate species. In developed countries, the ITS sequencing method can be a method of choice due to its high specificity, reliability, and shorter identification time. However, for a country like India, where disease registries, including fungal diseases, are currently unavailable, phenotypic identification, instead of the more expensive sequencing-based identification, can be reliably used to identify the common etiologic agents of clinical dermatophytosis.
ACKNOWLEDGMENTS
The author would like to acknowledge the Sri Balaji Vidyapeeth (Deemed to be University) for facilitating the conduct of this article.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors designed the experiments. NKB, RP and RM performed the experiments. All authors analyzed the data, wrote the manuscript and approved the manuscript for publication.
FUNDING
This work was supported by the SBV research grant number DeanRes/Res.grants/2019/563
ETHICS STATEMENT
The study was approved by the MGMC & RI Institutional Human Ethics Committee project number Faculty_Project/04/2019/06
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript. Universal ITS primer sequences are available from the corresponding author on reasonable request. ITS sequences have been deposited in GenBank with GenBank accession numbers MW454380, MW454772, MW454799, MW454828, MW454864, MW463449, MW463454, MW463455, MW463457, MW463460, MW463459, MW454821 and MW454827.
- Weitzman I, Summerbell RC. The dermatophytes. Clin Microbiol Rev. 1995;8(2):240-259.
Crossref - Cas ED. Parasitic adaptation of pathogenic fungi to mammalian hosts. Crit Rev Microbiol. 1986;13(2):173-218.
Crossref - King RD, Khan HA, Foye JC, Greenberg JH, Jones HE. Transferrin, iron, and dermatophytes. I. Serum dermatophyte inhibitory component definitively identified as unsaturated transferrin. J Lab Clin Med. 1975;86(2):204-212.
Crossref - Bristow IR, Spruce MC. Fungal foot infection, cellulitis and diabetes: A review. Diabet Med. 2009;26(5):548-551.
Crossref - Jerajani H, Janaki C, Kumar S, Phiske M. Comparative assessment of the efficacy and safety of sertaconazole (2%) cream versus terbinafine cream (1%) versus luliconazole (1%) cream in patients with dermtophytoses: a pilot study. Indian J Dermatol. 2013;58(1):34-38.
Crossref - Baldo A, Tabart J, Vermout S, et al. Secreted subtilisins of Microsporum canis are involved in adherence of arthroconidia to feline corneocytes. J Med Microbiol. 2008;57(9):1152-1156.
Crossref - Baldo A, Mathy A, Tabart J, et al. Secreted subtilisin Sub3 from Microsporum canis is required for adherence to but not for invasion of the epidermis. Br J Dermatol. 2010;162(5):990-997.
Crossref - Bagut ET, Baldo A, Mathy A, et al. Subtilisin Sub3 is involved in adherence of Microsporum canis to human and animal epidermis. Vet Microbiol. 2012;160(3-4):413-419.
Crossref - Baldo A, Chevigne A, Dumez ME, et al. Inhibition of the keratinolytic subtilisin protease Sub3 from Microsporum canis by its propeptide (proSub3) and evaluation of the capacity of proSub3 to inhibit fungal adherence to feline epidermis. Vet Microbiol. 2012;159(3-4):479-484.
Crossref - Liu T, Zhang Q, Wang L, et al. The use of global transcriptional analysis to reveal the biological and cellular events involved in distinct development phases of Trichophyton rubrum conidial germination. BMC Genomics. 2007;8:1-14.
Crossref - Grumbt M, Monod M, Yamada T, Hertweck C, Kunert J, Staib P. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J Invest Dermatol. 2013;133:1550-1555.
Crossref - Verma S, Vasani R, Reszke R, Matusiak L, Szepietowski JC. Prevalence and clinical characteristics of itch in epidemic- like scenario of dermatophytoses in India: a cross- sectional study. J Eur Acad Dermatol Venereol. 2020;34(1):180-183.
Crossref - Naglot A, Shrimali DD, Nath BK, et al. Recent trends of dermatophytosis in Northeast India (Assam) and interpretation with published studies. Int J Curr Microbiol Appl Sci. 2015;4:111-120.
- Rajagopalan M, Inamadar A, Mittal A, et al. Expert Consensus on The Management of Dermatophytosis in India (ECTODERM India). BMC Dermatol. 2018;18(1):6.
Crossref - Dogra S, Narang T. Emerging atypical and unusual presentations of dermatophytosis in India. Clin Dermatol Rev. 2017;1(3):12-18.
Crossref - Sahoo A, Mahajan R. Management of tinea corporis, tinea cruris, and tinea pedis: A comprehensive review. Indian Dermatol Online J. 2016;7(2):77-86.
Crossref - Dogra S, Uprety S. The menace of chronic and recurrent dermatophytosis in India: is the problem deeper than we perceive? Indian Dermatol Online J. 2016;7(2):73-76.
Crossref - Verma S, Madhu R. The great Indian epidemic of superficial dermatophytosis: an appraisal. Indian J Dermatol. 2017;62:227.
- Zhan P, Liu W. The Changing Face of Dermatophytic Infections Worldwide. Mycopathologia. 2017;182(1-2):77-86.
Crossref - Rebell G, Taplin D. Dermatophytes: Their recognition and identification, 2nd ed., University of Miami Press, Florida, 1970.
- Emmons C, Chapman H. Utz J. Culture media. In Medical Mycology, 2nd ed., Lea & Febiger, Philadelphia, 1971.
- Verrier J, Monod M. Diagnosis of dermatophytosis using molecular biology. Mycopathologia. 2017;182(1-2):193-202.
Crossref - Tartor YH, Abo Hashem ME, Enany S. Towards a rapid identification and a novel proteomic analysis for dermatophytes from human and animal dermatophytosis. Mycoses. 2019;62(12):1116-1126.
Crossref - Singh S, Beena PM. Comparative study of different microscopic techniques and culture media for the isolation of dermatophytes. Indian J Med Microbiol. 2003;21(1):21-24. PMID: 17642969
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes- application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113-118.
Crossref - Robert R, Pihet M. Conventional methods for the diagnosis of dermatophytosis. Mycopathologia. 2008;166(5-6):295-306.
Crossref - Graser Y, El Fari M, Vilgalys R, et al. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med Mycol. 1999;37(2):105-114.
Crossref - Graser Y, Kuijpers AFA, Presber W, De Hoog GS. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med Mycol. 1999;37(5):315-330.
Crossref - Frealle E, Rodrigue M, Gantois N, et al. Phylogenetic analysis of Trichophyton mentagrophytes human and animal isolates based on MnSOD and ITS sequence comparison. Microbiology. 2007;153(10):3466-3477.
Crossref - Elavarashi E, Kindo A, Kalyani J. Optimization of PCR-RFLP directly from the skin and nails in cases of dermatophytosis, targeting the ITS and the 18S ribosomal DNA regions. J Clin Diagnostic Res. 2013;7(4):646-651.
Crossref - Diongue K, Brechard L, Diallo MA, et al. A comparative study on phenotypic versus ITS-based molecular identification of dermatophytes isolated in Dakar, Senegal. Int J Microbiol. 2019;2019:6754058.
Crossref - Nasrin S, Saeed ZB, Sina M, Sahar N. Genotyping and molecular characterization of dermatophytes isolates collected from clinical samples. Arch Pulmonol Respir Care. 2017;3:52-57.
Crossref - Ahmadi B, Mirhendi H, Shidfar M, et al. A comparative study on morphological versus molecular identification of dermatophyte isolates. J Mycol Med. 2015;25(1):29-35.
Crossref - Neji S, Makni F, Sellami H, Cheikhrouhou F, Sellami A, Ayadi A. Molecular identification of dermatophytes isolated in Sfax-Tunisia. J Mycol Med. 2010;20(2):85-90.
Crossref - Venkatesan G, Singh AJAR, Murugesan AG, Janaki C, Shankar SG. Trichophyton rubrum the predominant etiological agent in human dermatophytoses in Chennai, India. Afr J Microbiol Res. 2007;1:9-12.
- Sumana V, Singaracharya MA. Dermatophytosis in Khammam (Khammam district, Andhra Pradesh, India). Indian J Pathol Microbiol. 2004;47(2):287-289.
- Bhatia VK, Sharma PC. Epidemiological studies on dermatophytosis in human patients in Himachal Pradesh, India. Springerplus. 2014;3:1-7.
Crossref - Kucheria M, Gupta SK, Chhina DK, Gupta V, Hans D, Singh K. Clinico-mycological profile of dermatophytic infections at a tertiary care hospital in North India. Int J Community Heal Med Res. 2016;2:17-22.
Crossref - Putta SD, Kulkarni VA, Bhadade AA, Kulkarni VN, Walawalkar AS. Prevalence of dermatophytosis and its spectrum in a tertiary care hospital, Kolhapur. Indian J Basic Appl Med Res. 2016;5:595-600.
- Ramaraj V, Vijayaraman RS, Rangarajan S, Kindo AJ. Incidence and prevalence of dermatophytosis in and around Chennai, Tamilnadu, India. Int J Res Med Sci. 2016;4(3):695-700.
Crossref - Pryce TM, Palladino S, Kay ID, Coombs GW. Rapid identification of fungi by sequencing the ITS1 and ITS2 regions using an automated capillary electrophoresis system. Med Mycol. 2003;41(5):369-381.
Crossref - Sharma M, Sharma R. Profile of dermatophytic and other fungal infections in Jaipur. Indian J Microbiol. 2012;52(2):270-274.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.