ISSN: 0973-7510
E-ISSN: 2581-690X
Carbapenems, frequently used for the treatment of infections caused by Multidrug-resistant (MDR) Gram-negative bacteria (GNB) are being reported with increased resistance rate. Colistin with other antibiotics has emerged as a saviour but inappropriate reporting of colistin susceptibility is a serious clinical concern. To detect the antimicrobial resistance of GNB isolates obtained from blood samples, further, colistin Minimum Inhibitory Concentration (MIC) testing of carbapenem-resistant isolates was done by the Broth Micro-dilution Method (BMD). This prospective study was done in the Department of Microbiology from January 2020 to December 2020. The GNB isolated from blood samples were identified & antimicrobial-susceptibility testing was performed by the Vitek 2 system. Colistin MIC of carbapenem-resistant isolates was done by the BMD method. The data were statistically analysed using SPSS 21. Growth was obtained in 5% of blood samples and 546 (75.8%) of these were GNB including predominantly E.coli, Klebsiella spp & Acinetobacter spp. Carbapenem-resistant infections 246 (48.5%) showed significant association with ICU admission, resistance to other classes of antibiotics & mortality. Among the carbapenem-resistant isolates, only seven (2.9%) were found resistant to colistin by the BMD test. Most of these were Klebsiella spp. (71.4%) & obtained predominantly from ICU patients (85.7%). All the carbapenem-resistant isolates were found intermediate sensitive to colistin by the VITEK-2 system. The isolates of GNB were characterized as MDR 323 (59.2%), XDR 164 (30%) and PDR 2 (0.4%). Use of, colistin, should be guided by BMD, the reference method for MIC testing to avoid erroneous reporting of colistin resistance.
Drug Resistance Multiple, Colistin, Carbapenems, Bloodstream Infections
Gram-negative bacterial infections resistant to multiple antibiotics have been increasing worldwide. Carbapenems are the broad-spectrum beta-lactam antibiotics frequently used for the treatment of these infections. According to WHO antibiotic resistance is among the top 10 threats to global public health while carbapenem-resistant Acinetobacter spp., Enterobacteriaceae & Pseudomonas spp., are listed as the top priority pathogens responsible for widespread concern.1 The commonest mechanism of resistance is by carbapenemases production which hydrolyze beta-lactam antibiotics including carbapenems. Approximately 70% of ICU infections are reported due to carbapenem-resistant GNB.2 Management of these infections has become a great challenge for clinicians due to the limited choice of antibiotics. Polymyxins and other antimicrobials such as tigecycline, carbapenems in high doses with aminoglycosides, and carbapenem double therapy are being used as potential treatment options.3 Polymyxins were first recognized in 1949 and used therapeutically for two decades. Later their use was discontinued due to side effects like nephrotoxicity & neurotoxicity. With a rise in the rates of carbapenem-resistant infections, it has re-emerged as the only therapeutic option in combination with other antibiotics,4-6 for MDR organisms. Polymyxin B and polymyxin E (colistin) are being used for treatment currently. Colistin targets lipopolysaccharides (LPS) in the outer membrane.7 There are sporadic reports of the emergence of colistin resistance from different parts of the world. Indian studies have reported varied prevalences of colistin resistance in Klebsiella pneumonia, Acinetobacter baumannii, and Pseudomonas aeruginosa ranging from 5% to 13%.4,7 In vitro susceptibility testing of colistin is challenging and is influenced by the multi-component composition of colistin; cationic property, adsorption to the microtiter plate, and heteroresistance to colistin. Considerable variations in the colistin AST results occurred due to discrepant MICs produced by gradient tests (E-test).4,7 Due to the inaccurate results of the commonly used methods, the use of disk diffusion, E-test & VITEK system are not recommended by CLSI (Clinical laboratory and standards institute) for colistin susceptibility testing while broth microdilution (BMD), and colistin agar dilution tests are the recommended methods.8,9 The therapeutic challenges include high nephrotoxicity and neurotoxicity, optimizing dosage, development of resistance during sub-optimal dosage, narrow therapeutic index, and lack of uniformity in dosing units with respect to critically ill patients.10
Underreporting of colistin-resistant bacteria is becoming a serious clinical concern. It is urgently needed that clinical microbiology laboratories identify accurate colistin susceptibility to avoid the use of this nephrotoxic antibiotic when it is not effective. Also, it will help to prevent the development of resistance to this valuable antibiotic. Most of the laboratories do not report colistin resistance by the reference method. Continuous surveillance of colistin resistance by the appropriate method is of utmost importance in the current situation. Hence this study was done to perform Gram-negative bacteria’s susceptibility to antibiotics is obtained from blood samples by the VITEK-2 system and further determination of colistin MIC of carbapenem-resistant Gram-negative isolates by Micro broth dilution technique.
This was a prospective, observational study conducted in the Department of Microbiology from January 2020 to December 2020 after the approval of the institutional ethics committee. The Gram-negative bacterial isolates obtained from blood samples received for culture & sensitivity from the patients admitted have been taken in the study.
Blood culture
The blood culture bottles were inoculated with 5-10 ml of blood from adult patients or 1-3ml of blood from paediatric patients and incubated in an automated system (BacT/ALERT, Biomerieux) system. Sub-cultures were done on blood agar and McConkey agar from positive culture bottles, and the growth obtained was further identified using Gram stain & the vitek2 system. Bottles were reported as culture-negative after 5 days of no growth.
Antimicrobial susceptibility testing (AST)
AST of the Gram-negative isolates was determined using the VITEK-2 system. The AST-GN405 card (Biomerieux) contained antibiotics amikacin, gentamicin, trimethoprim-sulfamethoxazole ciprofloxacin, cefepime, ceftriaxone, cefuroxime, amoxicillin-clavulanic acid, cefoperazone-sulbactam, piperacillin-tazobactam, imipenem, meropenem, and colistin was used for AST of lactose fermenting GNB.
The AST-GN406 card (Biomerieux) contained amikacin, gentamicin, trimethoprim-sulfamethoxazole, ciprofloxacin, cefepime, ceftazidime, cefotaxime cefoperazone/sulbactam, piperacillin/tazobactam, imipenem, meropenem, and colistin was used for AST of non-lactose fermenting GNB.
For quality check ATCC strains of Escherichia coli, 25922 and Pseudomonas aeruginosa 27853 were used. Results were noted as per CLSI (Clinical & Laboratory Standards Institute) guidelines. Carbapenem-resistant isolates were further tested by the Broth microdilution method. The organisms with intrinsic resistance to colistin like Burkholderia spp., Stenotrophomonas spp., Serratia spp., Proteus spp., Morganella spp. and Providencia spp., were excluded.
Broth microdilution method (BMD)
The test was conducted in accordance with CLSI guidelines. 1mg/ml stock solution of colistin sulphate (SIGMA-ALDRICH, 15000 units/mg) was prepared. Two-fold serial dilution of the 16ug/ml was done to achieve a concentration of 8ug/ml, 4ug/ml, and 2ug/ml, up to 0.25 µg/ml. A saline suspension of the isolate from an overnight incubated agar plate was prepared to obtain 0.5McFarland turbidity. A 25µl of each colistin dilution was added in a well-containing 25µl of organism suspension in 50ul of cation-adjusted Muller Hinton broth (CAMHB) media (2x strength) &. The total volume in each well of 96 well microtitre plates was 100µl. All the colistin dilutions were added in one column for one organism. Along with test columns, quality control columns consisted of bacterial control (inoculum + media), sterility control (drug + media), QC sensitive strain (E. coli ATCC 25922 + media + drug) & QC resistant strain ( mcr-1 strain NCTC 13846 Escherichia coli + media + drug) were tested simultaneously. The plate was incubated overnight at 37°C. The lowest concentration of antibiotic that completely inhibits the growth of the organism was noted as MIC. Results were interpreted as per CLSI guidelines.11 Enterobacterales /Pseudomonas/ Acinetobacter isolates with MIC ≤2 ug/ml were reported as intermediate sensitive and MIC ≥ 4 µg/ml were considered colistin-resistant.
The isolates were further characterized on the basis of antibiotic resistance as: –
MDR
The isolates showed resistance to at least one agent in three or more antimicrobial categories. Salmonella spp isolates resistant to ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole were considered as MDR.
XDR
The bacterial isolates showed resistance to at least one agent in all but two or fewer antimicrobial categories (i.e., susceptible to only one or two categories)
PDR
The isolates showed resistance to all agents in all antimicrobial categories and were noted as Pan Drug Resistant (PDR).
Statistical analysis
Data was analyzed as frequencies (number of cases), relative frequencies (percentage), range, and mean ± standard deviation. SPSS 21 version statistical program of Microsoft windows was used for statistical calculations. The Chi-Square test was used to calculate the P value and less than 0.05 was considered as significant.
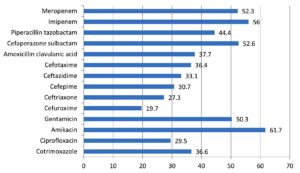
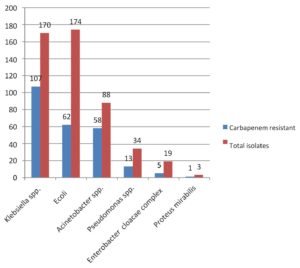
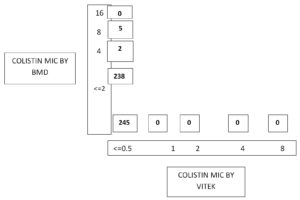
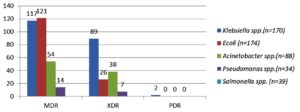
Out of 14,175 blood samples received for culture, 720 (5%) have shown growth. Among them, 546 (75.8%) Gram-negative bacterial isolates were isolated from the blood samples received from 364 male &182 female patients. A maximum of the patients with BSI belonged to the age group of 51-60 years (23.4%), followed by patients aged 0-10 years (15.7%). The enrolled patients’ average age was 45 and a half years. Among them, 365 (66.8%) were admitted to the wards & 181(33.2%) and to the ICUs. The distribution of these isolates is shown in Table 1. E. coli was the commonest GNB isolated, followed by Klebsiella spp & Acinetobacter spp. The antibiotic sensitivity profile of the GNB isolates is shown in Figure 1. Out of 546 GNB isolates, carbapenem susceptibility was interpreted for 507 isolates (Salmonella spp excluded). A total of 246 (48.5%) isolates were obtained from 172(70%) male patients & 74(30%) female patients were found resistant to carbapenems. The age range of these individuals with isolates that are resistant to carbapenem had been 48±2 years and the maximum samples were from the age group 41-60 years. Carbapenem resistance was seen significantly more in the patients admitted to the ICUs 97(55%) than to the wards149 (45%) (p<.05). The distribution of carbapenem-resistant resistant isolates is shown in Figure 2. AST profile of carbapenem sensitive and resistant isolates was compared (Table 2). A significant association was seen between carbapenem resistance and resistance to other classes of antibiotics. (P-value <0.05) Significantly high mortality (14%) was observed among the patients having BSI with carbapenem-resistant GNB. Out of carbapenem-resistant GNB isolates seven (2.9%) were found resistant to colistin by BMD test. Among the colistin-resistant isolates, mostly were Klebsiella spp. 5 (71.4%) followed by Acinetobater spp. 1(14.3%) & Psedomonas spp. 1(14.3%).(Table 3) Two of the Klebsiella spp isolates had MIC>4 ug/ml & other isolates had MIC of >8 ug/ml. (Figure 3) Colistin resistance was not seen in the other carbapenem-resistant isolates. While all the carbapenem-resistant isolates were found intermediate sensitive to colistin by the VITEK-2 system (MIC =<0.5 ug/ml). Among the seven patients with colistin-resistant infections, 6(85.7%) were admitted to the ICU. Of these two (28.5%) were discharged, two (28.5%) expired, and three (42.8%) were Discharged Against Medical Advice (DAMA). Out of 546 isolates, 223(40.8%) isolates were observed susceptible to most of the classes of antibiotics. The remaining isolates have shown resistance to many classes of antibiotics and were characterized as MDR 323 (59.2%), XDR 164 (30%) & PDR 2 (0.4%). (Figure 4).
Table (1):
Distribution of GNB isolates obtained from blood samples (n=546).
Gram negative isolates |
Number (%) |
|---|---|
Ecoli |
174(31.8) |
Klebsiella sp. |
170(31.2) |
Klebsiella pneumoniae Klebsiella oxytoca Klebsiella ozaenae |
164(30) 4(0.8) 2(0.4) |
Acinetobacter sp. |
88(16.1) |
Acinetobacter baumannii Acinetobacter lwoffii |
85(15.6) 3(0.5) |
Salmonella sp. |
39(6.9) |
Salmonella typhi Salmonella paratyphi A |
36(6.4) 3(0.5) |
Pseudomonas sp. |
34(6.3) |
Pseudomonas aeruginosa Pseudomonas stutzeri |
32(5.9) 2(0.4) |
Enterobacter cloacae complex |
19(3.5) |
Citrobacter koseri |
6(1.2) |
Achromobacter xylosans |
4(0.7) |
Stenotrophomonas maltophilia |
4(0.7) |
Proteus mirabilis |
3(0.6) |
Sphingomonas paucimobilis |
2(0.4) |
Burkholderia cepacia |
1(0.2) |
Serratia marcescens |
1(0.2) |
Elizabethkingia meningoseptica |
1(0.2) |
TOTAL |
546 |
Table (2):
Antimicrobial susceptibility profile of carbapenem sensitive and carbapenem resistant GNB isolates. (n=507).
| E. coli(n=174) |
Klebsiella spp (n=170) |
|||||
|---|---|---|---|---|---|---|
| Carbapenem Sensitive (n=112) | Carbapenem Resistant (n=62) | P-value | Carbapenem Sensitive (n=63) | Carbapenem Resistant (n=107) | P-value | |
| Cotrimoxazole | 54(48.2%) | 6(9.7%) | 0.001 | 50(79.4%) | 12(11.2%) | 0.001 |
| Ciprofloxacin | 33(29.4%) | 1(1.6%) | 0.001 | 46(73%) | 4(3.7%) | 0.001 |
| Amoxiclav | 63(56.3%) | 9(14.5%) | 0.001 | 51(80.9%) | 5(4.7%) | 0.001 |
| Amikacin | 109(97.3%) | 35(56.4%) | 0.001 | 62(98.4%) | 25(23.4%) | 0.001 |
| Gentamicin | 77(68.7%) | 20(32.2%) | 0.001 | 56(88.8%) | 19(17.7%) | 0.001 |
| Cefuroxime | 37(33%) | 3(4.8%) | 0.001 | 38(60.3%) | 4(3.7%) | 0.001 |
| Ceftriaxone | 36(32.1%) | 2(3.2%) | 0.001 | 37(58.7%) | 1(0.9%) | 0.001 |
| Cefepime | 51(45.5%) | 5(8.1%) | 0.001 | 40(63.4%) | 5(4.7%) | 0.001 |
| Cefaperazone sulbactam | 93(83%) | 11(17.7%) | 0.001 | 60(95.2%) | 5(4.7%) | 0.001 |
| Piperacillin tazobactam | 83(74.1%) | 10(16.1%) | 0.001 | 57(90.5%) | 5(4.7%) | 0.001 |
| Acinetobacter spp. (n=88) |
Pseudomonas spp. (n=34) |
|||||
| Carbapenem Sensitive (n=30) | Carbapenem Resistant (n=58) | P-value | Carbapenem Sensitive (n=21) | Carbapenem Resistant (n=13) | P-value | |
| Cotrimoxazole | 28(93.3%) | 5(8.6%) | 0.001 | 20(95.2%) | 0 | 0.001 |
| Ciprofloxacin | 29(96.7%) | 2(3.4%) | 0.001 | 17(80.9%) | 2(15.4%) | 0.001 |
| Amoxiclav | 30(100%) | 1(1.7%) | 0.001 | 20(95.2%) | 0 | 0.001 |
| Amikacin | 29(96.7%) | 3(5.2%) | 0.001 | 21(100%) | 4(30.7%) | 0.001 |
| Gentamicin | 30(100%) | 4(6.9%) | 0.001 | 21(100%) | 4(30.7%) | 0.001 |
| Cefepime | 24(80%) | 0 | 0.001 | 21(100%) | 7(53.8%) | 0.001 |
| Ceftazidime | 27(90%) | 0 | 0.001 | 20(95.2%) | 5(38.7%) | 0.001 |
| Cefotaxime | 29(96.7%) | 3(5.2%) | 0.001 | |||
| Cefaperazone sulbactam | 30(100%) | 14(24.1%) | 0.001 | 20(95.2%) | 7(53.8%) | 0.001 |
| Piperacillin tazobactam | 27(90%) | 0 | 0.001 | 20(95.2%) | 4(30.7%) | 0.001 |
Table (3):
Colistin susceptibility profile of Carbapenem resistant isolates by BMD method (n=245).
Organism |
Number of isolates n(%) |
Colistin intermediate isolates(MIC<=2ug/ml) n(%) |
Colistin resistant isolates (MIC>=4ug/ml) n(%) |
|---|---|---|---|
Klebsiella sp. |
107(43.7%) |
102(95.3%) |
5(4.7%) |
Ecoli |
62(25.3%) |
62(100%) |
0 (0%) |
Acinetobacter sp. |
58 (23.7%) |
57 (98.3%) |
1(1.7%) |
Pseudomonas sp. |
13(5.3%) |
12(92.3%) |
1(7.7%) |
Enterobacter cloacae complex |
5(2%) |
5(100%) |
0 (0%) |
Total |
245(100%) |
238(97.1%) |
7 (2.9%) |
Septicemia caused by MDR Gram-negative bacteria is a serious clinical concern in hospitals. In this prospective study, out of 720 BSI, 75.8% were due to Gram-negative bacteria. Many other studies have reported Gram-negative bacteria as the commonest cause of BSIs ranging between 60-80%.12,13 Similar to various other studies we reported bacteremia due to GNB predominantly in the male patients.12,14 In contrast Amipara R et al. have reported more infections among female patients.15 Like the few other studies, most of these infections observed in the patients belonged to the 51-60 years of age group (23.4%), followed by pediatric patients whose ages ranged from 0-10 years (15.7%) in our hospital, while, some of the other studies reported higher rates of BSI in the pediatric patients as compared to the adult patients.2,12 In this study common comorbidities observed among the patients with Gram-negative BSI were diabetes mellitus (22.3%), malignancy (10%), and hypertension (10%). Similarly, Diabetes mellitus followed by end-stage renal diseases were reported as the most common comorbidities among the patients with Gram-negative BSI in another study.15
Among the Gram-negative isolates, E. coli (31.8%) was the commonest isolate obtained in this study followed by Klebsiella sp (31.2%), Acinetobacter spp. (16.1%), Salmonella spp. (7.1%) and Pseudomonas spp. (6.2%). Leal HF et al. reported E. coli and K. pneumonia as the most frequently isolated pathogens followed by Pseudomonas aeroginosa. However, Palewar et al. have reported Acinetobacter spp. (14.6%) and Klebsiella spp. (14%) as the most common GNB isolated in their study.14,12
The AST pattern of E. coli, Klebsiella spp. and Acinetobacter spp. revealed high resistance to cotrimoxazole, ciprofloxacin, cephalosporins, and amoxicillin-clavulanic acid. E. coli isolates showed better sensitivity to piperacillin-tazobactam & cefoperazone-sulbactam as compared to Klebsiella spp. & Acinetobacter spp. isolates. Like the study of Zhang Z. et al., Pseudomonas isolates showed good susceptibility (60-80%) to most of the antibiotics.16 However, Susceptibility to aminoglycosides revealed encouraging results in all the GNB isolates which were in consistent with the study conducted by Bajaj A et al.13
Frequent use of carbapenems to treat the infections caused by the MDR organisms accounted for increasingly encountered resistance to these antibiotics. In our study, 246(48.5%) of the isolates were found resistant to carbapenems obtained predominantly from the blood samples of the male (70%) patients. The median age of the patients was 55 years. Similarly, the study by Porwal R et al. reported 64% carbapenem-resistant isolates from male patients and the mean age of the patients was 52.3 years.17 Consistent with the results of other studies, infections caused by carbapenem-resistant organisms were observed more in the patients admitted to the ICUs (55.1%) than in the wards.18,2 In the current study, Acinetobacter spp. (65.9%) and Klebsiella spp. (62.9%) showed higher resistance to carbapenems followed by Pseudomonas (38.2%). A study by Vamsi KS et al. showed 56% carbapenem resistance in Klebsiella spp. followed by Pseudomonas spp. (24%).2 Another study reported the highest carbapenem resistance in Acinetobacter spp. (53.5%).18 Similar to the other studies, we also observed, carbapenem-resistant isolates were significantly resistant to most of the other antibiotics (p-value <0.005).19 However, 56.4% of carbapenem-resistant E. coli isolates were found sensitive to amikacin and 54% of Pseudomonas spp. isolates were found sensitive to cefepime and cefoperazone sulbactam. Better aminoglycoside susceptibility among carbapenem-resistant isolates was reported by others also.7,18,19
Colistin has emerged as a ray of hope for the treatment of carbapenem-resistant GNB. In our study, seven (2.85%) carbapenem-resistant bacterial isolates were observed resistant to colistin. Out of these 85.7% were isolated from the samples of the ICU patients, Colistin resistant isolates were majorly seen in the patients who had prolonged hospital stays (>7 days). A hospital stay of >2 weeks was reported as a significant risk factor associated with colistin resistance.5 Further, it was noted that indwelling devices like endotracheal tubes, Catheters were present in all the patients harbouring colistin-resistant isolates. All of these patients had chronic comorbidities therefore frequent use of antibiotics in these patients could be the cause of resistance. Among all the colistin-resistant isolates five were Klebsiella spp. (71.4%) and one each of them was Acinetobacter spp. and Pseudomonas spp. Similarly, a study by Gupta P et al from Jaipur, Rajasthan reported five (5%) colistin-resistant isolates by the BMD method including two K. pneumonia, two P. aeruginosa as well as one A. baumannii. A few of the five resistant isolates, two isolates had a MIC of 4 µg/ml & three isolates had MIC of 8 µg/ml.4 A study by John P Mills reported 4 % of colistin-resistance among Enterobacterales in blood samples.20 A 6 years study from the USA, reported 128(0.45%) colistin-resistant Gram-negative isolates predominantly K. pneumoniae.21 Higher colistin resistance (14.9%) was reported from Vellore in the GNB with 20.6% and 9.1% resistance in Klebsiella spp. and Acinetobacter spp., respectively.22 Another study from Odisha reported 13.5% colistin resistance in CRE isolates.7 Very high (53%) colistin resistance was reported from Egypt.23 Lower colistin resistance in our study could be due to antibiotics usages in the hospital according to the antibiotic policy & stringent hospital infection control practices. All the colistin-resistant isolates by BMD were found intermediate sensitive to colistin by the Vitek-2 system. Hence, this system cannot be relied on for colistin susceptibility testing. It should be confirmed by the BMD method which is recommended by CLSI. like us, Khurana S et al. observed that the automated Vitek 2 method could not detect the resistance in up to 48.5% of GNB isolates in their study. When compared both test results, 88% categorical agreement was observed.6 In contrast, Shaikh S et al. have noted that the Vitek method showed resistance but these isolates were found susceptible by the BMD method & the isolates had a discrepancy in the MIC values by both methods while Gupta P et al. have reported a high level of agreement with the reference BMD method.24,4 Limitation of our study was molecular detection of colistin resistance could not be performed due to the unavailability of resources.
In our study MDR (59.2%), XDR (30%), PDR (0.4%) isolates were observed and the maximum XDR isolates were Klebsiella spp. followed by Acinetobacter spp. All the PDR isolates were identified as Klebsiella spp. In the study by Basak S et al., 37% MDR, 14% XDR and none of the PDR isolate was observed.25
We observed more deaths (14%) among the patients harbouring infection with carbapenem-resistant GNB and high mortality in patients who had colistin-resistant infections (28.6%). DAMA patients were terminally ill; if included in the death, mortality reached 45% among patients with carbapenem-resistant infections and 71.4% in patients who had colistin-resistant infections. Other studies have reported 40-60% mortality in patients infected with colistin-resistant bacteria.26,27
Treatment of carbapenem-resistant infections is challenging as they are resistant to most of the classes of antibiotics. Usually, the laboratories do not perform the colistin susceptibility (MIC) test by the appropriate method. Antibiotic surveillance of colistin-resistant infections and use of the last-resort antibiotic only after MIC determination by BMD is needed as under-reporting of the colistin-resistant bacteria is an important reason for misuse of this nephrotoxic antibiotic.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Dayanand Medical College and Hospital, Ludhiana, India, with reference number DMCH/P/2019/1758.
- Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15.
Crossref - Vamsi KS, Ramamoorthy S, Murali TS, Vamsi A, Reddy BR, Hameliamma M. Prevalence of Carbapenem Resistant Gram Negative Bacteria in Rural Hospital Mahabubnagar, Telangana and Systemic Review. Int J Curr Microbiol App Sci. 2021;10(3):1542-1547.
Crossref - Ruppe E, Woerther PL, Barbier F Mechanisms of antimicrobial resistance in Gram-negative bacilli Ann Intensive Care. 2015;5:61.
Crossref - Gupta P, Sharma R, Vyas A, Tak A. Comparative evaluation of broth microdilution with E-test, Vitek 2, and disk diffusion for susceptibility testing of colistin on Gram-negative bacteria. Indian J Med Sci. 2021;73(1):93-98.
Crossref - Kaza P, Mahindroo J, Veeraraghavan B, Mavuduru RS, Mohan B, Taneja N. Evaluation of risk factors for colistin resistance among uropathogenic isolates of Escherichia coli and Klebsiella pneumoniae: a case-control study. J Med Microbiol. 2019;68(6):837-847.
Crossref - Khurana S, Malhotra R, Mathur P. Evaluation of Vitek®2 performance for colistin susceptibility testing for Gram-negative isolates. JAC Antimicrob Resist. 2020;2(4):101.
Crossref - Kar P, Behera B, Mohanty S, Jena J, Mahapatra A. Detection of Colistin Resistance in Carbapenem Resistant Enterobacteriaceae by Reference Broth Microdilution and Comparative Evaluation of Three Other Methods. J Lab Physicians. 2021;13(3):263-269.
Crossref - Bakthavatchalam YD, Veeraraghavan B. Challenges, issues, and warnings from CLSI and EUCAST working group on polymyxin susceptibility testing. J Clin Diagn Res. 2017;11(8):DL03-DL04.
Crossref - Singhal L, Sharma M, Verma S, et al. Comparative evaluation of broth microdilution with polystyrene and glass-coated plates, agar dilution, E-Test, Vitek, and disk diffusion for susceptibility testing of colistin and polymyxin B on carbapenem-resistant clinical isolates of Acinetobacter baumannii. Microb Drug Resist. 2018;24(8):1082-1088.
Crossref - Velkov T, Roberts KD, Thompson PE, Li J. Polymyxins: a new hope in combating Gram-negative superbugs? Future Med Chem. 2016;8(10):1017-1025.
Crossref - CLSI. Clinical and Laboratory Standard Institute. Performance standards for antimicrobial susceptibility testing. CLSI supplement M100. 29th edition, CLSI, Wayne, PA, 2021.
- Palewar M, Mudshingkar S, Dohe V, Kagal A, Karyakarte R. Bacteriological profile and antibiogram of blood culture isolates from a tertiary care hospital of Western India. J Datta Meghe Inst Med Sci Univ. 2020;15(2):261-265.
Crossref - Bajaj A, Mishra B, Loomba PS, et al. Prevalence of Gram-negative Septicemia in a Tertiary Care Center. J Med Sci Health. 2019;5(1):36-41.
Crossref - Leal HF, Azevedo J, Silva GE, et al. Bloodstream infections caused by multidrug-resistant gram-negative bacteria: epidemiological, clinical and microbiological features. BMC Infect Dis. 2019;19:1-1.
Crossref - Amipara R, Winders HR, Justo JA, Bookstaver PB, Kohn J, Al- Hasan MN. Impact of follow up blood cultures on outcomes of patients with community-onset gram-negative bloodstream infection. E Clinical Medicine. 2021;34:100811.
Crossref - Zhang Z, Chen M, Yu Y, Pan S, Liu Y. Antimicrobial susceptibility among gram-positive and gram-negative blood-borne pathogens collected between 2012-2016 as part of the Tigecycline Evaluation and Surveillance Trial. Antimicrob Resist Infect Control. 2018;7:152.
Crossref - Porwal R, Gopalakrishnan R, Rajesh NJ, Ramasubramanian V. Carbapenem resistant Gram-negative bacteremia in an Indian intensive care unit: A review of the clinical profile and treatment outcome of 50 patients. Indian J Crit Care Med. 2014;18(11):750-753.
Crossref - Xu A, Zheng B, Xu YC, Huang ZG, Zhong NS, Zhuo C. National epidemiology of carbapenem-resistant and extensively drug-resistant Gram-negative bacteria isolated from blood samples in China in 2013. Clin Microbiol Infect. 2016;22(Suppl 1):S1-S8.
Crossref - Mohanty S, Gajanand M, Gaind R. Identification of carbapenemase-mediated resistance among Enterobacteriaceae blood stream isolates: A molecular study from India. Indian J Med Microbiol. 2017;35(3):421-425.
Crossref - Mills JP, Rojas LJ, Marshall SH, et al. Risk Factors for and Mechanisms of Colistin Resistance Among Enterobacterales: Getting at the CORE of the Issue. Open Forum Infect Dis. 2021;8(7):ofab145.
Crossref - Richter SE, Miller L, Uslan DZ, et al. Risk Factors for Colistin Resistance among Gram-Negative Rods and Klebsiella pneumoniae Isolates. J Clin Microbiol. 2018;56(9):e00149-18.
Crossref - Bakthavatchalam YD, Shankar A, Thukaram B, Krishnan DN, Veeraraghavan B. Comparative evaluation of susceptibility testing methods for colistin and polymyxin B among clinical isolates of carbapenem- resistant Klebsiella pneumoniae and Acinetobacter baumannii. J Infect DevCtries. 2018;12(6):504-507.
Crossref - Anan MMG, El-Seidi EA, Mostafa MS, Rashed LA, El-Wakil DM. Detection of Plasmid-Mediated Mobile Colistin Resistance Gene (mcr-1) in Enterobacterales Isolates from a University Hospital. Infect Drug Resist. 2021;14:3063-3070.
Crossref - Shaikh S, Pandya H, Arora S, Kamtekar T. Comparison of Colistin Susceptibility Testing by Vitek 2 Compact and Broth Microdilution Method for Carbapenem Resistant Isolates in a Tertiary Diagnostic Centre. Int J Intern Med Geriatr. 2019; 1:54-58.
- Basak S, Singh P, Rajurkar M. Multidrug Resistant and Extensively Drug Resistant Bacteria: A Study. J Pathog. 2016;2016:4065603.
Crossref - Capone A, Giannella M, Fortini D, et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect. 2013;19(1):E23-E30.
Crossref - Prawang A, Santimaleeworagun W, Changpradub D, Thunyaharn S, Puttilerpong C. Treatment and Clinical Outcomes Among Infected Patients with Colistin-resistant Klebsiella pneumoniae Bacteremia. Open Forum Infectious Diseases. 2019;6(Suppl. 2):S782-S783.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.