ISSN: 0973-7510
E-ISSN: 2581-690X
Resistant strains of Klebsiella pneumonia are one of the most prevalent cause of nosocomial and especially respiratory tract infections al-around the world. The present investigation was carried out to study the prevalence of antibiotic resistance in K. pneumonia strains of hospitalized patients suffered from RTIs. Three-hundred and fifty respiratory samples were collected from hospitalized patients of both sexes and various ages. Samples were cultured and those that were K. pneumonia-positive were subjected to 16S rRNA based-PCR amplification and disk diffusion method. Of 350 samples studied, 25 samples were positive for K. pneumonia (7.14%). Distribution of K. pneumonia in male and female patients were 8.33% and 5.88%, respectively (P <0.05). Older than 60 years old and younger than 20 years old patients had the highest prevalence of K. pneumonia. Our K. pneumonia isolates had the highest levels of resistance against gentamycin (96%), ampicillin (92%), erythromycin (84%), ciprofloxacin (76%), sulfamethoxazole (76%). Primary identification of K. pneumonia-positive patients and their treatment with imipenem antibiotic based on the results of disk diffusion method can control distribution of K. pneumonia RTIs.
Klebsiella pneumonia, Prevalence, Antibiotic resistance, Respiratory Tract Infections, Risk factors.
Respiratory tract infections (RTIs) are one of the most common types of infectious diseases al-around the world. It has been estimated that more than 15% of death in developed countries are occurred due to the occurrence of RTIs1,2. RTIs are responsible for about 55,000 cases of infection and hospitalization in 20101, 2. They are accounted for more than 45,000 hospital admissions with an average length of stay of 6.3 days1.
In despite of a lot of developments in treatment and control of nosocomial infections, Klebsiella pneumonia (K. pneumonia) infections are remain as one of the most prevalent cause of morbidity and mortality al-around the world (3-5). It is a Gram-negative, rod-shaped, non-motile, encapsulated, lactose-fermenting, facultative anaerobic bacterium and widely distributed in the urinary, gastrointestinal, and respiratory tracts of healthy people. K. pneumonia is a causative agent of opportunistic infections mainly nosocomial and hospital-acquired infections including RTIs, pneumonia, Urinary Tract Infections (UTIs), abscesses, wound infection, diarrhea, sepsis and inflammation3-5.
In addition to the invasive nature of K. pneumonia, clinical strains of this bacterium are mainly resist to several types of antibiotic agents6-8. Antibiotic resistant strains of K. pneumonia cause more sever and dangerous infections for longer periods of time than susceptible strains (6-8). It has been estimated that K. pneumonia strains of RTIs and also other types of nosocomial infections had a high prevalence of antibiotic resistance (30-100%) against commonly used antimicrobial agents including ²-lactam antibiotics, broad-spectrum penicillins, cephalosporins, , carbapenems, fluoroquinolones, aminoglycosides, and tetracyclines6-9.
According to the uncertain status of K. pneumonia RTIs in Iran and lack of epidemiological investigations in the field of antibiotic resistance, the present investigation was carried out to study the prevalence and antibiotic resistance pattern of K. pneumonia strains isolated from the respiratory samples of male and female patients of various age groups hospitalized due to the severe RTIs in Iranian hospitals.
Samples collection
From January to November 2015, a total of 350 respiratory samples including Broncho Alveolar Lavages (BAL) (n=50) and also respiratory secretions (n=300) were sent to our laboratory center from hospitalized patients suffering from RTIs. In this study, all of respiratory samples were selected and analyzed for presence of L. pneumophila. At the time of sampling, information about the age, sex and clinical symptoms of the patients were recorded. Ten ml of each sample was immediately transferred to a sterile falcon tube containing ice and was immediately transferred to the laboratory.
Isolation of Klebsiella pneumoniae
All samples were transferred to the laboratory using peptone water (BPW, Merck, Germany). Samples were transferred to the McConkey agar (MC agar, Merck, Germany), Chocolate agar (Merck, Germany and Blood agar (Merck, Germany). The inoculum on the plate was streaked out for discrete colonies with a sterile wire loop. The culture plates were incubated at 37°C for 24 hours and observed for growth through the formation of colonies. All the bacteria were isolated and identified using morphological, microscopy of standard methods (API System; bioMerieux, Marcy-l’Étoile, France) and biochemical tests including Triple Sugar Iron agar (TSI agar, Merck, Germany), Simmons Citrate, Methyl Red, Indole, Urease, Voges–Proskauer, motility, and H2S production. The biochemical characteristics of K. pneumoniae identified were as follows: positive citrate utilization test, negative methyl red test, negative indole test, positive urease test, positive Voges–Proskauer test, sucrose, acid and abundant gas production from glucose, lactose, mannitol sugar fermentation tests, and maltose.
DNA extraction and PCR confirmation of Klebsiella pneumonia isolates
Bacterial isolates were cultured another time on nutrient broth media (Merck, Germany) and incubated aerobically at 370C for 24 h. Genomic DNA was extracted from each K. pneumoniae isolate by DNA extraction kit (Fermentas, Germany) according to manufacturer’s instruction. The bacteria were confirmed using the PCR method for 16S rRNA gene of the K. pneumonia (10). PCR was carried out with 2 µL template DNA, 0.25 µM of each primer (F: 5’-GCAAGTCGAGCGGTAGCACAG-3’ and R: 5’-CAGTGTGGCTGGTCATCCTCTC-3’) (260 bp), 0.2 mM deoxyribonucleoside triphosphates, 1x reaction buffer, 2 mM MgCl2 and 1.5 U Taq DNA polymerase (Fermentas) in a total volume of 25 µL. The DNA was amplified using the following protocol: initial denaturation (95 ºC for 5 min), followed by 30 cycles of denaturation (95ºC for 45 s), annealing (95ºC for 45 s) and extension (72ºC for 1 min), with a single final extension of 5 min at 72ºC. The programmable thermal cycler (Eppendorf, Mastercycler® 5330, Eppendorf-Netheler-Hinz GmbH, Hamburg, Germany) PCR device was used in all PCR reactions. Ten microliters of PCR products were resolved on a 1.5% agarose gel containing 0.5 mg/ml of ethidium bromide in Tris–borate–EDTA buffer at 90 V for 1 h, also using suitable molecular weight markers. The products were examined under ultraviolet illumination.
Antibiotic susceptibility test
K. pneumoniae isolates were incubated initially on the nutrient agar media (at 4°C) and their positive colonies were transferred to the Mueller– Hinton agar (Merck, Germany). Antimicrobial susceptibility was performed on Mueller-Hinton agar by the standard disk diffusion method recommended by the Clinical and Laboratory Standards Institute.11 This was done by dipping a sterile swab stick in to overnight nutrient broth and carefully swabbing the entire surface of Mueller–Hinton agar plates. Susceptibility of K. pneumoniae strains were determined against tetracycline (30 µg/disk), ceftazidime (30 µg/disk), ciprofloxacin (5 µg/disk), sulfamethoxazol (25 µg/disk), ampicillin (10 u/disk), trimethoprim (5 µg/disk), gentamycin (10 µg/disk), ceftriaxone (30 u/disk), amikacin (10 µg/disk), imipenem (30 µg/disk), erythromycin (15 µg/disk), and amoxicillin/clavulanic acid (20/10 µg/disc) antibiotic disk (Oxoid, UK). Plates were incubated at 370C for 18–24 h. The diameter of zone of inhibition was measured in millimeters and isolates were scored as sensitive or resistant by comparing with values recommended on standard charts (11). K. pneumoniae ATCC 43816 was used as quality control organism in antimicrobial susceptibility determination.
Statistical analysis
The results were transferred to a Microsoft Excel spreadsheet (Microsoft Corp., Redmond, WA) for analysis. Statistical analysis was performed using SPSS/16.0 software (SPSS Inc., Chicago, IL) for significant relationship between incidence and pattern of resistance of K. pneumoniae strains of RTIs. Chi-square and Fisher’s exact 2-tailed tests were performed in this study. Statistical significance was regarded at a P value < 0.05.
Table 1 represents the total prevalence of K. pneumonia in various studied groups. Twenty-five out of 350 samples (7.14%) were positive for presence of K. pneumonia. On the other hand, total prevalence of K. pneumonia in male and female patients of our study were 8.33% and 5.88%, respectively. Results of the culture method were also confirmed by the 16S rRNA gene-based PCR amplification. Figure 1 shows results of the gel electrophoresis for the PCR amplification of 16S rRNA gene. Among all studied groups, older than 60 years old male patients (10%) and also younger than 20 years old female patients (12%) had the highest prevalence of bacteria. Statistically significant differences were seen for the prevalence of K. pneumonia between various age groups (P <0.05).
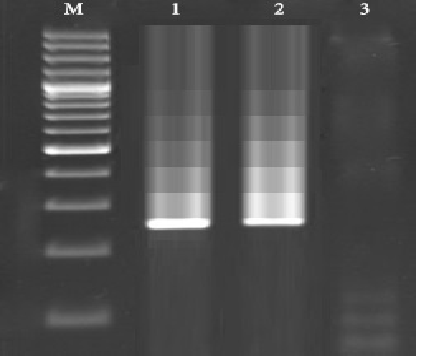 Figure 1. Results of the gel electrophoresis for amplification of 16s rRNA gene of the K. pneumonia isolated from samples of hospitalized patients with severe RTIs. M: 100 bp ladder (Fermentas, Germany), 1: Positive sample for 16s rRNA gene of the K. pneumonia (260 bp), 2: Positive control (K. pneumoniae ATCC 43816) and 3: Negative control (sterile distilled water (Merck, Germany).
Figure 1. Results of the gel electrophoresis for amplification of 16s rRNA gene of the K. pneumonia isolated from samples of hospitalized patients with severe RTIs. M: 100 bp ladder (Fermentas, Germany), 1: Positive sample for 16s rRNA gene of the K. pneumonia (260 bp), 2: Positive control (K. pneumoniae ATCC 43816) and 3: Negative control (sterile distilled water (Merck, Germany). Table (1):
Distribution of K. pneumonia in the samples taken from patients suffered from RTIs.
| Samples of RTIs | No. samples collected | K. pneumonia positive samples (%) | PCR confirmation of K. pneumonia (%) | |
|---|---|---|---|---|
| Male | <20 | 20 | 2 (10) | 2 (10) |
| 20-40 | 30 | 2 (6.66) | 2 (6.66) | |
| 4-60 | 50 | 3 (6) | 3 (6) | |
| >60 | 80 | 8 (10) | 8 (10) | |
| Total | 180 | 15 (8.33) | 15 (8.33) | |
| Female | <20 | 25 | 3 (12) | 3 (12) |
| 20-40 | 25 | 1 (4) | 1 (4) | |
| 4-60 | 50 | 2 (4) | 2 (4) | |
| >60 | 70 | 4 (5.71) | 4 (5.71) | |
| Total | 170 | 10 (5.88) | 10 (5.88) | |
| Total | 350 | 25 (7.14) | 25 (7.14) | |
Table 2 represents the antibiotic resistance pattern of K. pneumonia strains of patients suffered from RTIs. We found that K. pneumonia strains of our study harbored the highest levels of resistance against gentamycin (96%), ampicillin (92%), erythromycin (84%), ciprofloxacin (76%), sulfamethoxazole (76%) and amoxicillin/clavulanic acid (82.28%). The lowest levels of resistance were seen against imipenem (8%). Bacterial strains which were isolated from male patients had the higher prevalence of resistance than those of females (P <0.05).
Table (2):
Antibiotic resistance pattern of K. pneumonia in the samples taken from patients suffered from RTIs.
| Samples (No. K. pneumonia positive) | Antibiotic resistance pattern (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tet30 | cftz | cip5 | sul25 | am10 | tri5 | gen10 | cftx | amik | imp | eryt | amclv | |
| Male (15) | 10 (66.66) | 10 (66.66) | 12 (80) | 12 (80) | 14 (93.33) | 13 (86.66) | 15 (100) | 11 (73.33) | 13 (86.66) | 2 (13.33) | 12 (80) | 11 (73.33) |
| Female (10) | 3 (30) | 4 (40) | 7 (70) | 7 (70) | 9 (90) | 5 (50) | 9 (90) | 3 (30) | 4 (40) | – | 9 (90) | 7 (70) |
| Total (25) | 13 (52) | 14 (65) | 19 (76) | 19 (76) | 23 (92) | 18 (72) | 24 (96) | 14 (56) | 17 (68) | 2 (8) | 21 (84) | 18 (72) |
Tet30: tetracycline (30 µg/disk), cftz: ceftazidime (30 µg/disk), cip5: ciprofloxacin (5 µg/disk), sul25: sulfamethoxazol (25 µg/disk), am10: ampicillin (10 u/disk), tri5: trimethoprim (5 µg/disk), gen10: gentamycin (10 µg/disk), cftx: ceftriaxone (30 u/disk), amik: amikacin (10 µg/disk), imp: imipenem (30 µg/disk), eryt: erythromycin (15 µg/disk), and amclv: amoxicillin/clavulanic acid (20/10 µg/disc)
The present investigation was focused on the study the prevalence rate and antimicrobial resistance pattern of K. pneumonia strains isolated from the samples taken from patients suffered from RTIs. We found that 7.14% of samples were positive for K. pneumonia with higher prevalence in males and also older than 60 and younger than 20 years old patients. One possible explanation for the high prevalence of K. pneumonia in our study is the fact that all of the tested samples were taken in the winter season. Coldness can effect on the prevalence of RTIs and is an important predisposing factor for occurrence of pneumonia and RTIs caused by pathogenic agents like K. pneumonia.
A conceivable explanation for the higher prevalence of K. pneumonia in male than female is that men usually have more contact with the contaminated external environment. They work outside the house but women usually stay at home and are not in close contact with outside. In fact, most of the Iranian women prefer to work at home. In addition, higher levels of immunity in men than women caused to their infection with resistant strains of K. pneumonia. Older patients had also the lower levels of immunity and therefore they had a higher prevalence of K. pneumonia. The reason for the higher prevalence of K. pneumonia in younger than 20 years old patients is the fact that the population of this group of patients of our study were infants and pediatrics suffered from pneumonia and RTIs. Therefore, based on their lower levels of immunity, occurrence of RTIs caused by K. pneumonia were high.
Irregular and unauthorized prescription of antibiotics is the main factor for such high prevalence of antibiotic resistance in the K. pneumonia strains of our study. According to this thesis, it is not surprising that K. pneumonia strains of our investigation harbored the high levels of resistance against tetracycline, ceftazidime, ciprofloxacin, sulfamethoxazol, ampicillin, trimethoprim, gentamycin, ceftriaxone, amikacin, imipenem, erythromycin and amoxicillin/clavulanic acid. Higher prevalence of antibiotic resistance in isolates of male than female is due to their higher levels of immunity. Therefore, they only be infected with resistant strains of K. pneumonia.
Several investigations have been conducted in this field al-around the world. Jafari Sales et al. (2015)12 revealed the high prevalence of K. pneumonia in the hospital infections of Iranian hospitals with considerable levels of resistance against et al ceftazidime (100%), cefotaxime (98.60%), ciprofloxacin (82%), amikacin (53.30%) and gentamicin (47.60%) which was in harmony with our findings. Sanchez et al. (2013)6 reported increasing prevalence of antibiotic resistance in the K. pneumonia strains of clinical infections from 1998 to 2010. They showed that the prevalence of resistance in the American isolates of K. pneumonia in 2010 against tetracycline. amikacin, imipenem, gentamycin, cefepime, trimethoprim/sulfamethoxazole, piperacillin/tazobactam, tobramycin, ceftriaxone, ciprofloxacin, ceftazidime and aztreonam were 16.70%, 4.50%, 4.30%, 9.20%, 7.70%, 19.30%, 12.70%, 13.80%, 12.10%, 16.80%, 17.20% and 22.20%, respectively which was lower than our results in 2016. Kalaskar et al. (2012)13 reported that the prevalence of resistance in the K. pneumonia strains of clinical infections against ampicillin, amoxicillin/clavulanate, gentamycin, amikacin, cefuroxime, ciprofloxacin, trimethoprim/sulfamethaxole and imipenem were 100%, 100%, 100%, 37.50%, 22%, 88.60%, 86.80%, 10% and 0%, respectively which was similar with our results. Similar results have been reported from India (2011)14, Australia (2015)15, Switzerland (2016)16, South Africa (2012)17 and Brazil (2015)18.
Availability of antibiotic agents, cost of them and idea of medical practitioners for prescription of antibiotics are the main factors caused differences in the prevalence of antibiotic resistance in various studies.
The present investigation is one of the widespread reports of prevalence and antibiotic resistance pattern of K. pneumonia isolated from samples of hospitalized patients suffered from RTIs. The most important findings of our study are higher prevalence of K. pneumonia in males and also older than 60 years old and younger than 20 years old patients and also inefficiency of gentamycin, ampicillin, erythromycin, ciprofloxacin, sulfamethoxazole and amoxicillin/clavulanic acid prescription. It seems that primary identification of sources of K. pneumonia RTIs and their treatment with regular and accurate prescription of imipenem, according to the results of disk diffusion method can control distribution of K. pneumonia RTIs. Study the pattern of antibiotic resistance not only for K. pneumonia but also for other types of human clinical infections is necessary for each province/states, city and even each hospital.
- Bettering the Evaluation and Care of Health (BEACH) study: URTI / bronchosinusitis in general practice. Australian Institute of Health and Welfare General Practitioner Statistics and Classification Centre, University of Sydney [Westmead Hospital’s Family Medicine Research Centre]. March 2005.
- Hoyert DL, Xu J. Deaths: Preliminary Data for 2011. Natl Vital Stat Rep. 2012; 61(6):1-51.
- Lou Z, Qi Y, Qian X, Yang W, Wei Z. Emergence of Klebsiella pneumoniae carbapenemase-producing Escherichia coli sequence type 131 in Hangzhou, China. Chin Med J (Engl). 2014; 127(3):528-31.
- Darvishi M. Virulence Factors Profile and Antimicrobial Resistance of Acinetobacter baumannii Strains Isolated from Various Infections Recovered from Immunosuppressive Patients. Biomedical & Pharmacology Journal. 2016; 3:1057-1062.
- Sadeta Hadi, Amer ustovi, Jasmina Smajlovi, Sead Ahmetagi. Distribution of nosocomial infections caused by Klebsiella pneumoniae ESBL strain. J environ Occup Sci 2012; 1(3): 141-146.
- Sanchez GV, Master RN, Clark RB, Fyyaz M, Duvvuri P, Ekta G, Bordon J. Klebsiella pneumoniae antimicrobial drug resistance, United States, 1998-2010. Emerg Infect Dis. 2013; 19(1):133-6.
- Koksal F, Ak K, Kucukbasmaci O, Samasti M. Prevalence and antimicrobial resistance patterns of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from blood cultures in an Istanbul University Hospital. Chemotherapy. 2009; 55(4): 293-7.
- Mansury D, Motamedifar M, Sarvari J, Shirazi B, Khaledi A. Antibiotic susceptibility pattern and identification of extended spectrum b-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae from Shiraz, Iran. Iran J Microbiol. 2016; 8(1):55-61.
- Feizabadi MM, Etemadi G, Yadegarinia D, Rahmati M, Shabanpoor S, Bokaei S. Antibiotic-resistance patterns and frequency of extended-spectrum beta-lactamase-producing isolates of Klebsiella pneumoniae in Tehran. Med Sci Monit. 2006; 12(11):BR362-5.
- Tayebeh F, Amani J, Moradyar M, Mirhossaini SA, Detection of Klebsiella Pneumoniae 16s rDNA Specific Gene by PCR-ELISA Technique. Journal of Fasa University of Medical Sciences 2016; 5: 545-550.
- Clinical and Laboratory Standards Institute (CLSI): Performance standards for antimicrobial susceptibility testing. 22nd informational supplement. M100–S22. Wayne (PA): The Institute; 2012.
- Jafari Sales A, Jafari B, Beygoli N. Antimicrobial Resistance Patterns in Extended-spectrum ²lactamase Producing Klebsiella pneumoniae Isolates in a Razi Hospital Marand, Iran. Electronic Journal of Biology, 2015, 11(1): 8-12
- Kalaskar A, Venkataramana K. Determination of Antimicrobial Resistance Pattern and Production of Extended-Spectrum b-Lactamases amongst Escherichia coli and Klebsiella pneumoniae from Clinical Isolates. J Med Bacteriol. 2012, 1: 17-24
- Singh Sikarwar A, Vardhan Batra H. Prevalence of Antimicrobial Drug Resistance of Klebsiella pneumoniae in India. International Journal of Bioscience, Biochemistry and Bioinformatics. 2011; 1: 211-215.
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H,Wilksch J, Gorrie C, Schultz MB, Edwards DJ1, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C,Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015; 112(27):E3574-81.
- Jayol A, Nordmann P, Desroches M, Decousser JW, Poirel L. Acquisition of Broad-Spectrum Cephalosporin Resistance Leading to Colistin Resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2016; 60(5):3199-201.
- Perovic O, Singh-Moodley A, Dusé A, Bamford C, Elliott G, Swe-Han KS, Kularatne R, Lowman W, Whitelaw A, Nana T, Wadula J, Lekalakala R,Saif A, Fortuin De-Smit M, Marais E. National sentinel site surveillance for antimicrobial resistance in Klebsiella pneumoniae isolates in South Africa, 2010 – 2012. S Afr Med J. 2014 Jun 19; 104(8):563-8.
- Prestes-Carneiro LE, Azevedo AM, Nakashima MA, Xavier JM, Cabral C. Frequency and Antimicrobial susceptibility of pathogens at tertiary public hospital, Sao Paulo, Brazil. Southeast Asian J Trop Med Public Health. 2015 Mar; 46(2):276-84.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


