ISSN: 0973-7510
E-ISSN: 2581-690X
Vachellia tortilis is a tree belonging to the family Fabaceae that inhabits high temperature and dry environments of the world. It is ecologically and economically significant amongst others of the genera Vachellia and Senegalia. It is native to Namibia and several other countries. Various parts of the V. tortilis plant are used for different therapeutic purposes both in the traditional and pharmacological settings. However, V. tortilis is vulnerable to pathogenic infection to which they lack natural resistance and little is known regarding the pathogens responsible for such infections. The aim of the study was to isolate and identify the fungal species associated with twig dieback disease in V. tortilis. Pure cultures from diseased V. tortilis were grown using potato dextrose agar (PDA) and DNA was subsequently isolated and later amplified in a PCR reaction using ITS1 and ITS4 primers. Sequencing and BLAST search revealed the identity of the isolates as; Penicillium rubefaciens, Penicillium herbarum, Trichoderma longibrachiatum and Trichoderma harzianum. Results indicated that P. herbarum was associated with disease symptoms in A. tortilis, T. longibrachiatum and T. harzianum were antagonistic fungi while the effect of P. rubefaciens on V. tortilis remained unknown. It is recommended that further investigations using Koch’s postulates should be performed on V. tortilis using the isolates.
Penicillium rubefaciens, Penicillium herbarum, Trichoderma longibrachiatum, Trichoderma harzianum, Namibia, Vachellia tortilis.
Acacia, Vachellia and Senegalia species largely inhabit high temperature and dry environments of Africa. A total of about 1200 species of of these genera have been identified and are prominent in Arabia, Australia and Africa (Baldwin et al., 1999; Yadav et al., 2013). Acacia, Vachellia and Senegalia species are of economic importance because they are used to produce various materials such as tannin, gum and timber (Ibrahim and Aref, 2000; Yadav et al., 2013). They also provide valuable fodder for livestock and are also ecologically important in the ecosystems within they occur.
Vachellia tortilis is a tree that grows up to 21 m tall belonging to the family Fabaceae. It is a significant species amongst others of the genus Vachellia and is native to Namibia, Angola, Botswana, Egypt, Eritrea, Ethiopia, Iran, Israel, Kenya, India, Pakistan, Mozambique, Qatar, Saudi Arabia, Somalia, South Africa, Sudan, Swaziland, Tanzania, Uganda, United Arab Emirates, Zambia and Zimbabwe (Wickens et al, 1995). V. tortilis contains important properties that are beneficial commercially and medicinally. Various parts of the V. tortilis plant are used for different therapeutic purposes both in the traditional and pharmacological settings.
Traditionally, V. tortilis plant parts are described to have the following uses; seed for food, leaves and fruits for fodder, flower for forage, wood for fuel and timber, bark for dyestuff (tanning), gum as a food additive, and whole plant for soil fertility (Fterich et al., 2012; Anderson, 1993; Baldwin et al., 1999; Abdallah et al., 2008; Oba, 1998; Satya and Jindal, 1994; Yadav et al., 2013). Pharmaceutical uses of the V. tortilis plant include; polysaccharide isolated from gum exudates for diabetes mellitus, stem bark for fungal diseases and infectious diseases (mouth and dental), bark tannins for diarrhea, wood for dry cough, root for cough and diphtheria, root bark for malaria, aqueous extract for hyper cholesterol and inflammation, and methanol extracts for leishmanial and parasitic diseases (Abdallah and Merito, 2013; Bisht et al., 2013; Kigondu et al., 2009; Maregesi et al., 2008; Maregesi et al., 2007; Alharbi and Azmat, 2011; Nguta and Mbaria, 2013; Njoroge and Bussmann, 2006; Tunon et al., 1995).
Although V. tortilis has a wide variety of uses, it remains vulnerable to infection from an increasing number of pathogens to which they lack natural resistance. Information regarding the entire pathogens associated with disease in V. tortilis will provide a basis for assessing the condition of the disease and develop appropriate breeding programs to minimise losses. Hence, the main aim of the study was to isolate and identify the fungal species associated with twig dieback disease in V. tortilis.
Isolation of fungi
A piece of diseased V. tortilis stem was surface-sterilized by submerging in 99% ethanol for 7 minutes and rinsed five times with sterile distilled water. The surface sterilized stem was dissected in a laminar flow and pieces of its pith were transferred using flamed tweezers onto potato dextrose agar (PDA) for growth. In addition, white basidiomycete buds that were attached to the periderm of the diseased V. tortilis bark were plucked with flamed tweezers and placed onto potato dextrose agar (PDA) and left to grow. The PDA plates were then incubated at 35±2°C for 14 days. After which subcultures were created by aseptically transferring distinct fungi growing from the V. tortilis plant parts on the PDA media onto fresh PDA media and incubated for 10 days at 35±2°C.
DNA extraction
Fungal genomic DNA was extracted using the Zymo Research Fungal/Bacterial DNA MiniPrep™ kit according to the manufacturer’s protocol. The extraction was performed on one week old fungal cultures and the DNA was stored at 4°C for later use as the template for amplification. After successful isolation, the DNA (7µl) was mixed with 2µl of 6X loading dye and run on a 1.5% agarose gel prepared in 1X TAE buffer and viewed using UV illumination.
PCR amplification and Sequence analysis
The DNA was amplified in 50 µl PCR reaction volumes using ITS-1 (52 -TCCGTAGGTGAACCTGCGG-32) and ITS-4 (52 -TCCTCCGCTTATTGATATGC-32) primers according to White et al. (1990) with minor modifications. The PCR reaction was carried out in a Bio-Rad MyCycler™ thermal cycler and the mixture contained 4µl of template DNA, 2µl of a 0.5µM concentration of ITS1 primer, 2µl of a 0.5µM concentration of ITS4 primer, 17µl of nuclease free water and 25 µl of 2x Dream Taq master mix which contained: Dream Taq DNA polymerase, 2x Dream Taq buffer, dATP, dCTP, dGTP and dTTP of 0.4mM each, and 0.4 mM MgCl2. The PCR reaction profile consisted of initial denaturation temperature of 94ºC for 4 min, followed by 40 cycles of denaturation temperature at 95ºC for 1 min, annealing temperature of 55ºC for 1 min, and an extension temperature at 72ºC for 45 seconds. The final extension was then performed at 72ºC for 10 min and lastly the PCR products were held at 4ºC. The products were separated by electrophoresis on a 2% agarose gel stained with ethidium bromide and visualized under UV light.
The PCR fragments were purified and sequenced at Inqaba Biotechnical Industries (Pty) Ltd (Pretoria, South Africa). The obtained sequences were then edited in BioEdit (Biological Sequence Alignment editor for Windows 99/98/NT/2K/XP/7) sequence alignment editor software (Hall, 1998). BLAST searches were then perform in the NCBI GenBank database and the sequences possessing the highest similarity with the query sequence were chosen reflecting the identity of the isolates in question.
The culturing of V. tortilis sections of the stem and bark resulted in the growth of four morphologically distinct pure colonies (fig.1 and fig. 2). PCR was performed using ITS1 and ITS4 primers and electrophoresis results showed expected ITS amplicons of about 1450 bp.
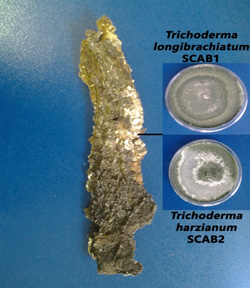 Fig. 1. White basidiomycete from diseased A. tortilis bark grown on PDA media
Fig. 1. White basidiomycete from diseased A. tortilis bark grown on PDA media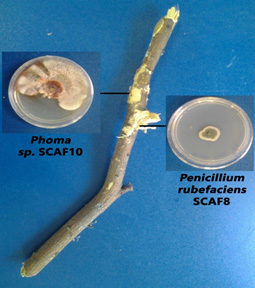 Fig. 2. Fungal isolates from diseased A. tortilis stem grown on PDA media
Fig. 2. Fungal isolates from diseased A. tortilis stem grown on PDA mediaA BLAST search using the A. tortilis ITS sequences revealed the identities of the isolates as; Penicillium rubefaciens, Phoma herbarum, Trichoderma longibrachiatum and Trichoderma harzianum. The isolates were obtained from the A. tortilis as follows; P. rubefaciens from the Acacia pith, P. herbarum from the Acacia bark, and T. longibrachiatum and T. harzianum from the basidiomycetes on the bark.
Table (1):
The bacteria isolates and their corresponding identity retrieved from NCBI.
Fungal isolate |
Plant Host |
NCBI Identity |
% Identity |
Accession Number |
|---|---|---|---|---|
F8 |
V. tortilis |
Penicillium rubefaciens |
100 |
KR905616.1 |
F10 |
V. tortilis |
Phoma herbarum |
99 |
KP144996.1 |
B1 |
V. tortilis |
Trichoderma longibrachiatum |
100 |
KP326575.1 |
B2 |
V. tortilis |
Trichoderma harzianum |
99 |
LN714612.1 |
The results revealed the identity of the isolates from the diseased V. tortilis as P. rubefaciens, P. herbarum, T. longibrachiatum and T. harzianum. All these fungi have not been reported previously on this plant species in Namibia. Penicillium and other fungus such as Aspergillus and Talaromyces (Eurotiomycetes) are amongst the genera that are commonly found indoors (Amend et al. 2010; Houbraken et al. 2011).
Penicillium species have been reported (Frisvad et al. 2004; Frisvad and Samson 2004) to be linked to bio deterioration in foods especially fruits and producing various mycotoxins. Penicillium species have also been positively used in the food industry for the production of cheese (Giraud et al. 2010; Visagie et al., 2014), fermented sausages (Lopez-Díaz et al., 2001; Ludemann et al., 2010), and medical industry for the production of penicillin (Visagie et al., 2014). Penicillium, Eurotium and Cladosporium species have also been isolated from the cork (outer layer of the Quercus suber L. tree.) by Barreto et al. (2012) and they argued that these fungal species colonize the cork alongside different kinds of fungi inhabiting different parts of the tree and surrounding soil. Depending on the fungal species, some form a parasitic relationship while others form a saprobiotic one in which case the cork provides the relevant nutrients for the fungal growth (Barreto et al., 2012). Moreover, Penicillium occurs in diverse environments that include soil, vegetation, air, indoor environments and in food (Visagie et al., 2014). To our knowledge, this is the first report on the presence of P. rubefaciens in A. tortilis. The effect of P. rubefaciens on A. tortilis remains unknown and further investigations to assess if this species is the primary disease causing pathogen on A. tortilis are vital.
The Phoma genus is diverse and composed of species that inhabit a wide range of environments such as soil and plants. A substantial amount of the species of this genus are saprophytic with some being opportunistic (Irinyi et al., 2007). In addition, most coelomycetes are associated with plants and in some cases are primary pathogens (Sutton, 1980). According to Boerema et al. (2005), approximately 2000 species of Phoma have been isolated and characterized. Furthermore, Garas et al. (2012) labeled Cytosperma chrysoperma and Phoma species such as; P. glomerata, P. cava and P. eupyrena as pathogens responsible for dieback disease in V. mellifera and V. karoo.
P. herbarum species have been isolated from fruit trees in which they infect the leaves and twigs (Valiuskaite, 2002) and sapwood of healthy and declining scots pine trees (Giordano et al., 2009). Giordano et al. (2009) revealed that P. herbarum, Thanatephorus cocumeris and Mucor plumbeus species are associated with the existence of mistletoes on scots pine trees. A mistletoe is a parasitic plant that affects the stand structure by inducing crown losses and tree mortality but also has some ecological importance in the survival of some trees because it functions as a carbon sink (Cullings et al., 2005; Mueller and Gehring, 2006). Giordano et al. (2009) further argued that mistletoe on a tree influences the diversity of endophytic mycoflora inhabiting that particular tree and declining scots pine trees possess different endophytic mycobiota in their sapwood compared to that of healthy trees. However, P. herbarum has been designated as the primary pathogen leading to twig dieback in A. mearnsii after its consistent isolation from diseased A. mearnsii trees (Gibson, 1979; Olembo, 1972; Roux et al., 1995). Gibson (1979), Olembo (1972) and Roux et al. (1995) further argued that P. herbarum caused fresh infections through wounds. The present study did not observe the presence of mistletoe on the V. tortilis tree but can infer that P. herbarum is associated with disease in the V. tortilis.
Trichoderma species are widely found in the soil and root ecosystems and are used as biological control agents against a variety of plant pathogens especially fungi (Mulaw et al., 2013). These species are found in a diverse range of habitats ranging from soil and growing on wood, bark, and other fungi and numerous substrates (Druzhinina et al., 2011). This report is the first one on Trichoderma occurring in V. tortilis in Namibia. The commonly used Trichoderma species as biological controls are T. harzianum, T. atroviride, T. viride, T. virens and T. koningii. These species are antagonistic microorganisms capable of preventing the growth of pathogens in the soil and plant (Singh et al., 2014). However, the effect of Trichoderma species on non-target organisms is less understood although it is relevant for their effective use. Trichoderma species have found prominent use because they prevent increase of the pathogens, produce enzymes that degrade the cell wall of pathogens, produce antibiotics capable of destroying pathogens, induce the development of the plant, and induce plant defensive mechanisms (Singh et al., 2014). Druzhinina et al. (2011) explained that the antagonistic properties of parasitizing and killing other fungi are common in Trichoderma species and have thus earned them widespread use as biological control organisms. Studies have revealed that T. longibrachiatum and T. harzianum are used as biological agents against fungal infection in plants (B³aszczyk et al., 2014; Howell, 2003; Limon et al., 2004; Mohd Zainudin et al., 2008; Mulaw et al., 2013).
The isolation of T. longibrachiatum and T. harzianum in A. tortilis in the present study suggests the relationship between the A. tortilis, and T. longibrachiatum and T. harzianum species is either commensal or mutualistic because of their antagonistic properties. It was revealed that P. herbarum is associated to twig dieback disease in V. tortilis while the effect of P. rubefaciens on V. tortilis remains unknown. In addition, pathogens have the ability to form synergistic relationships which have been detected in various plant species (Lamichhane and Venturi, 2015). Synergistic relationships range from fungi-fungi interaction, bacteria-bacteria interaction, virus-virus interaction, and mixed interactions such as fungi-bacteria interaction. Synergism justifies the existence of plant diseases in which more than a single pathogen is involved in the infection process (Lamichhane and Venturi, 2015). Synergism and its effects go undetected in monocultures and it probably occurs frequently in plant diseases. Glen et al. (2009) revealed fungi-fungi synergism in A. mangium infected with root rot disease in which the pathogens responsible were Ganoderma philippii, Ganoderma mastoporum, Ganoderma austral, and Amauroder marugosum. Consequently, it is possible that some of the isolates in this study were opportunistic pathogens that benefit from synergism. Moreover, with the current data set, it is not possible to draw firm conclusions that P. herbarum is the primary cause of the twig dieback although it is associated with twig dieback as reported in previous studies (Gibson, 1975; Olembo, 1972; Roux et al., 1995) because of the possible existence of other fungal species that are not or rarely detected in culture based methods.
The study aimed at isolating and identifying the culturable fungal species associated with twig dieback disease in V. tortilis in which case four fungal species were successfully isolated and identified as; P. rubefaciens, P. herbarum, T. longibrachiatum and T. harzianum. Of the four isolates, P. herbarum was detected as the specie associated with the observed disease in A. tortilis. T. longibrachiatum and T. harzianum were identified as antagonistic fungi with the ability to protect the V. tortilis tree against pathogenic fungi but with the potential to be pathogenic depending on the circumstances. However, the effect of P. rubefaciens on V. tortilis remained unknown. It is therefore recommended that further; investigations using Koch’s postulated should be performed on V. tortilis using the isolates, studies should be performed using more informative community analysis molecular techniques that are able to detect the entire community and circumvent the limitations of culture based methods; and microbial synergistic relationships leading to disease in V. tortilis should be investigated in order to deduce whether the twig dieback disease is caused by a single primary pathogen or it is a result of complex interactions.
ACKNOWLEDGMENTS
This study was supported by the research grant of the Research and Publications Office at the University of Namibia grant URPC/2015/221.
- Abdallah, F., Noumi, Z., Touzard, B., Belgacem, A.O., Neffati, M. and Chaieb, M., The influence of Acacia tortilis (Forssk.) subsp. raddiana (Savi) and livestock grazing on grass species composition, yield and soil nutrients in arid environments of South Tunisia. Flora-Morphology, Distribution, Functional Ecology of Plants, 2008; 203(2), pp.116-125.
- Abdallah, H. A. and Merito, A., Medicinal plants and their uses by the people in the Region of Randa, Djibouti. Journal of Ethnopharmacology., 2013; 148: pp.701–713.
- Alharbi, W.D.M. and Azmat, A.I.S.H.A., Hypoglycemic and hypocholesterolemic effects of Acacia tortilis (Fabaceae) growing in Makkah. Pakistan Journal of Pharmacology, 2011; 28(1): pp.1-8.
- Amend, A.S., Seifert, K.A., Samson, R. and Bruns, T.D., Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proceedings of the National Academy of Sciences, 2010; 107(31), pp.13748-13753.
- Anderson D.M.W., Some factors influencing the demand for gum arabic (Acacia Senegal (L.) and other water soluble tree exudates. Forest Ecology and Management., 1993; 58: pp.1-18.
- Baldwin, T. C., Quah, P. E., Menzies, A. R., A serotaxonomic study of Acacia gum exudates. Phytochemistry, 1999; 50: pp.599-606.
- Barreto, M.C., Houbraken, J., Samson, R.A., Brito, D., Gadanho, M. and San Romao, M.V., Unveiling the fungal mycobiota present throughout the cork stopper manufacturing process. FEMS microbiology ecology, 2012; 82(1), pp.202-214.
- Bisht, S., Kant, R. and Kumar, V., á-d-Glucosidase inhibitory activity of polysaccharide isolated from Acacia tortilis gum exudate. International journal of biological macromolecules, 2013; 59, pp.214-220.
- B³aszczyk, L., Siwulski, M., Sobieralski, K., Lisiecka, J. and Jêdryczka, M., Trichoderma spp.–application and prospects for use in organic farming and industry. Journal of Plant Protection Research, 2014; 54(4), pp.309-317.
- Boerema, G. H., Gruyter, J., Noordeloos, M. E., and Hamers M. E. C., Phoma identification manual. CABI Publishing. CAB International Wallingford, Oxfordshire, UK. 470 pp. Novenyvedelem, 2004; 41(4), pp.172-173.
- Cullings, K., Raleigh, C. and Vogler, D.R., Effects of severe dwarf mistletoe infection on the ectomycorrhizal community of a Pinus contorta stand in Yellow stone Park. Canadian Journal of Botany, 2005; 83, pp. 1174-1180.
- Druzhinina, I.S., Seidl-Seiboth, V., Herrera-Estrella, A., Horwitz, B.A., Kenerley, C.M., Monte, E., Mukherjee, P.K., Zeilinger, S., Grigoriev, I.V. and Kubicek, C.P., Trichoderma: the genomics of opportunistic success. Nature Reviews Microbiology, 2011; 9(10), pp.749-759.
- Frisvad, J. and Samson, R. A., Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology, 2004; 49, pp. 1–174.
- Frisvad, J.C., Smedsgaard, J., Larsen, T.O. and Samson, R.A., Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Studies in Mycology, 2004; 49, pp.201-241.
- Fterich, A., Mahdhi, M. and Mars, M., Impact of grazing on soil microbial communities along a chronosequence of Acacia tortilis subsp. raddiana in arid soils in Tunisia. European Journal of Soil Biology, 2012; 50, pp.56-63.
- Garas, L. S, Uzabakiriho, J. D and Chimwamurombe, P. M., Isolation and Identification of Fungal Species Associated with Gall Formation on Acacia mellifera in Western Windhoek. Journal of Pure and Applied Microbiology, 2012; 6(2), pp. 713-716.
- Gibson, I.A.S., Diseases of forest trees widely planted as exotics in the tropics and southern hemisphere. Part II. The genus Pinus. Commonwealth Forestry Institute 1979.
- Giordano, L., Gonthier, P., Varese, G.C., Miserere, L. and Nicolotti, G., Mycobiota inhabiting sapwood of healthy and declining Scots pine (Pinus sylvestris L.) trees in the Alps. Fungal Diversity, 2009; 38, pp. 69-83.
- Giraud, F., Giraud, T., Aguileta, G., Fournier, E., Samson, R., Cruaud, C., Lacoste, S., Ropars, J., Tellier, A. and Dupont, J., Microsatellite loci to recognize species for the cheese starter and contaminating strains associated with cheese manufacturing. International journal of food microbiology, 2010; 137(2), pp.204-213.
- Glen, M., Bougher, N.L., Francis, A.A., Nigg, S.Q., Lee, S.S., Irianto, R., Barry, K.M., Beadle, C.L. and Mohammed, C.L., Ganoderma and Amauroderma species associated with root-rot disease of Acacia mangium plantation trees in Indonesia and Malaysia. Australasian Plant Pathology, 2009; 38(4), pp.345-356.
- Hall, T. BioEdit. Biological sequence alignment editor for Windows. North Carolina, USA: Carolina State University 1998.
- Houbraken, J., Frisvad, J.C. and Samson, R.A., Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus: The Global Mycological Journal, 2011; 2(1), pp.87.
- Howell, C.R., Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Disease, 2003; 87(1), pp.4-10.
- Ibrahim, A.A. and Aref, I.M., Host status of thirteen Acacia species to Meloidogyne javanica. Journal of nematology, 2000; 32(4S), pp.609.
- Irinyi, L.M., Kövics, G. and Karaffa, E.M., Classification of Phoma species using new phylogenetic marker 2007.
- Kigondu, E. V. M., Rukunga, G. M. and Keriko, J. M., Anti-parasitic activity and cytotoxicity of selected medicinal plants from Kenya. Journal of Ethno pharmacology. 2009; 123, pp. 504–509.
- Lamichhane, J.R. and Venturi, V., Synergisms between microbial pathogens in plant disease complexes: a growing trend. Frontiers in Plant Science, 2015; 6, pp.385.
- Limon, M.C. and Codón, A.C., Biocontrol mechanisms of Trichoderma strains. International Microbiology, 2004; 7, pp.249-260.
- López-Dýµaz, T.M., Santos, J.A., Garcýµa-López, M.L. and Otero, A., Surface mycoflora of a Spanish fermented meat sausage and toxigenicity of Penicillium isolates. International Journal of Food Microbiology, 2001; 68(1), pp.69-74.
- Ludemann, V., Greco, M., Rodríguez, M.P., Basílico, J.C. and Pardo, A.G., Conidial production by Penicillium nalgiovense for use as starter cultures in dry fermented sausages by solid state fermentation. LWT-Food Science and Technology, 2010; 43(2), pp.315-318.
- Maregesi, S.M., Ngassapa, O.D., Pieters, L. and Vlietinck, A.J., Ethnopharmacological survey of the Bunda district, Tanzania: Plants used to treat infectious diseases. Journal of Ethnopharmacology, 2007; 113(3), pp.457-470.
- Maregesi, S.M., Pieters, L., Ngassapa, O.D., Apers, S., Vingerhoets, R., Cos, P., Berghe, D.A.V. and Vlietinck, A.J., Screening of some Tanzanian medicinal plants from Bunda district for antibacterial, antifungal and antiviral activities. Journal of Ethnopharmacology, 2008; 119(1), pp.58-66.
- Mohd Zainudin, N.A.I. and Abdullah, F., Disease suppression in Ganoderma-infected oil palm seedlings treated with Trichoderma harzianum. Plant Protection Science, 2008; 44(3), pp.101-107.
- Mueller, R.C. and Gehring, C.A., Interactions between an above ground plant parasite and below ground ectomycorrhizal fungal communities on pinyon pine. Journal of Ecology, 2006; 94(2), pp.276-284.
- Mulaw, T.B., Druzhinina, I.S., Kubicek, C.P. and Atanasova, L., Novel endophytic Trichoderma spp. isolated from healthy Coffea arabica roots are capable of controlling coffee tracheomycosis. Diversity, 2013; 5(4), pp.750-766.
- Nguta J.M. and Mbaria J.M., Brine shrimp toxicity and antimalarial activity of some plants traditionally used in treatment of malaria in Msambweni district of Kenya. Journal of Ethnopharmacology 2013; 148, pp. 988–992.
- Njoroge, G.N. and Bussmann, R. W., Herbal usage and informant consensus in ethno veterinary management of cattle diseases among the Kikuyus (Central Kenya). Journal of Ethno pharmacology, 2006; 108, pp. 332–339.
- Oba, G., Effects of excluding goat herbivory on Acacia tortilis woodland around pastoralist settlements in northwest Kenya. Acta Oecologica, 1998; 19(4), pp.395-404.
- Olembo, T.W., Phoma herbarum Westend.: A pathogen of Acacia mearnsii De Wild. in Kenya. East African Agricultural Forest Journal. 1972.
- Roux, J., Kemp, G.H.J. and Wingfield, M.J., Diseases of black wattle in South Africa—A review. South African Forestry Journal, 1995; 174(1), pp.35-40.
- Singh, A., Shahid, M. and Srivastava, M., Phylogenetic relationship of Trichoderma asperellum Tasp/8940 using Internal Transcribed Spacer (ITS) sequences. International Journal, 2014; 2(3), pp.979-986.
- Sutton, B.C., The Coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. 1st edn. Commonwealth Mycological Institute, U.K 1980.
- Tunon, H., Olavsdotter, C. and Bohlin, L., Evaluation of anti-inflammatory activity of some Swedish medicinal plants. Inhibition of prostaglandin biosynthesis and PAF-induced exocytosis. Journal of Ethnopharmacology, 1995; 48(2), pp.61-76.
- Valiuskaite, A., Micromycetes infecting stone fruit trees. Biologija, 2002; 1: pp.18-21.
- Vir, S. and Jindal, S.K., Fruit infestation of Acacia tortilis (Forsk) hyne by Bruchidius andrewesi Pic.(Coleoptera: Bruchidae) in the Thar desert. Forest ecology and management, 1994; 70(1), pp.349-352.
- Visagie, C.M., Houbraken, J., Frisvad, J.C., Hong, S.B., Klaassen, C.H.W., Perrone, G., Seifert, K.A., Varga, J., Yaguchi, T. and Samson, R.A.. Identification and nomenclature of the genus Penicillium. Studies in Mycology, 2014; 78: pp.343-371.
- White, T.J., Bruns, T., Lee, S.J.W.T. and Taylor, J.W., Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: A guide to methods and applications, 1990; 18: pp.315-322.
- Wickens, G.E., Sief El Din, A.G., Sita, G. and Nahal, I., Role of Acacia species in the rural economy of dry Africa and the Near East (No. 27). 1995.
- Yadav, P., Kant, R. and Kothiyal P., A Review on Acacia tortilis. International Journal of Pharmaceutical and Phytopharmacological Research, 2013; 3(2), pp. 93-96.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


