ISSN: 0973-7510
E-ISSN: 2581-690X
MicroRNAs (miRNAs) are short non-coding RNAs consist of 20-24 nucleotides, it play major roles in all biological pathways in mammals and other multicellular organisms. it effected in numerous cancer-relevant processes like proliferation, cell cycle control, programed cell death, differentiation, migration and metabolism. The present study carried out to estimate Micr-RNA146a gene polymorphism in breast cancer tissue, about 30 sample of embedded tissue were collected after diagnostic it, also 30 sample of healthy blood were collected for comparison , micro-RNA gene was detected using PCR-SSCP technique, the results show that there were 4 haplotypes in this gene A,B,C,D . The Variations in these haplotypes appeared in A and D, haplotypes A and was appeared in 75%, and 100% in patients and control respectively, D disappeared in control but it was 100% in patients . The present study concluded that this gene can be conducted as risk factor in breast cancer, but it need more investigations to detect SNPs and sequencing of micro-RNA146a gene.
Micr-RNA146a, gene polymorphism, breast cancer, PCR-SSCP.
Cancer is primarily diagnosed by clinical presentation, radiology, biochemical tests, and pathological analysis of tumor tissue, increasingly supported by molecular diagnostic tests. Molecular profiling of tumor tissue samples has emerged as a potential cancer classifying method (Akbani et al., 2014; Han et al., 2014; Hoadley et al., 2014).
microRNA (miRNA) is of interest as a regulatory mechanism in genomic expression. The human genome project identified approximately 20,000 genes in humans, but only 2 % of these genes are translated into proteins (International Human Genome Sequencing Consortium, 2004). A large-scale transcriptome analysis showed that RNAs are also transcribed in several noncoding regions, in which gene sequences of proteins are not encoded. These so-called non-coding RNAs (ncRNAs) have complex functions including regulation of chromatin structure, splicing, and protein silencing. miRNA is the most important class of ncRNAs. miRNAs are small RNA molecules of about 22 nucleotides that induce gene silencing. About 2,000 miRNAs have been found in humans and these molecules are important for biological activities including development; cell differentiation, proliferation and death; and metabolism ( Elbashir et al., 2001; Stefani and Slack, 2008).
Various diseases are caused by breakdown of gene regulatory networks due to changes in miRNA levels, and the relationship of miRNAs with cancer is under investigation. One miRNA regulates many messenger RNAs (mRNAs), while cancer does not develop due to a single cause, but due to breakdown of multiple mechanisms. Therefore, in contrast to common drugs, miRNAs may be used as treatment through comprehensive regulation of expression of molecules related to cancer. Use of miRNAs may also contribute to elucidation of oncogenic and homeostatic mechanisms.
miRNA-induced RNA silencing is involved in homeostatic maintenance of organisms and breakdown of this mechanism causes disease. Cancer is among the best studied diseases caused by failure of miRNA regulation. miRNAs are implicated in cancer development and progression, and expression patterns of miRNAs in normal tissues differ from those of cancer tissues. miRNAs involved in cancer are classified into oncogenic miRNAs (oncomiRs), which are up regulated in tumor tissues; and tumor suppressor miRNAs (tumor suppressor miRs), which are down regulated in tumor tissues and inhibit cancer. OncomiRs target mRNAs that suppress tumor growth, whereas tumor suppressor miRs target mRNAs that promote tumor growth. Failed regulation of both types of miRNAs promotes oncogenesis, proliferation and invasion of cancer cells, and the epithelial-mesenchymal transition (EMT).
Involvement of miRNAs in human cancer was first shown in B cell chronic lymphocytic leukemia (CLL), in which expression of miR-15 or miR-16 was found to decrease (Calin et al., 2002). In cancers, miR- 17-92 is up regulated in lung cancer cells (Hayashita et al., 2005). A large-scale analysis of miRNA expression in 540 patients with lung, breast, gastric, prostate, colon and pancreatic cancer showed that expression profiles of miRNAs in cancer tissues differ from those in normal tissues (Volinia et al., 2006). miR-21 was particularly up regulated in all types of cancer, indicating that it is likely to be involved in cancer onset. Subsequently, miR-21 was shown to regulate transcriptional repressors including NFIB (Fujita et al., 2008). Up regulation of miR-21 down regulates transcriptional repressors, resulting in transcriptional activation; therefore, miR- 21 is considered to be a typical oncomiR. miRNAs that are down regulated in cancer have also been found. For example, let-7, which was found in C. Elegans and is involved in cell growth, is a typical miRNA precursor that is down regulated in breast, lung and gastric cancer. let-7 targets oncogenes including RAS and high mobility group AT-hook 2 (HMGA2) and has been shown to be a tumor suppressor miR (Johnson et al., 2005; Lee and Dutta, 2007; Yu et al., 2007). Some miRNAs have carcinoma specific differential expression. The relationship of many of these miRNAs with their target genes is unknown, but miR-19 is necessary for inhibition of apoptosis and oncogenesis in lymphocytes (Olive et al., 2009). Chen et al. (2007) found that undifferentiated cell clusters including embryonic stem (ES) cells, embryoid bodies and mouse 11-day embryos had expression profiles of miRNAs that were simpler than those of mature somatic cells. These results suggest that the differentiation stage may be defined by profiles of specific miRNAs. It is also of note that miRNA profiles in undifferentiated stem cells are similar to those of cancer cells.
Sample collection
About 30 breast cancer embedded tissue was collect from histopathology unit in Al-Saader medical city, these samples was diagnosis by specialist physician as a breast tumor tissue, also all samples were used to diagnosis tumors in females that don’t treated with any anticancer therapy, and 30 blood sample of control were collected from healthy female that have age (35-65 years).
DNA extraction and PCR
DNA was extracted from embedded tissue according to the leaflet of Geneaid manufacture with modification according to Al-Terehi et al., (2016), in briefly : About 40 mg of tissue section was put in eppendorf tube contain 1 ml of xylene, it mixed and incubate at room temperature for 15 min, then it Centrifuged at 14000 rpm for3 min, the supernatant removed, Absolute ethanol was added (1 ml) to mixture then it Centrifuged at 14000 for3 min, supernatant was removed, , GT buffer was added (200µl) with homogenize by micropestle, then Proteinase K (40 µl) was added and incubate for 20min. at 60 C with inverting every 5 min. GBT buffer was added (200 µl) with mixing, then incubate at 60 C for 20 min and Absolute ethanol was added (200 µl) with mixing, then transfer mixture to GD column after that it Centrifuged at 14000 rpm for 2 min, the flow-through was discarded and W1 buffer was added to column (400 µl), then Centrifugation at 14000rpm for 20 sec. the flow-through was discarded also. Wash buffer was added (600 µl) Centrifuged columns at 14000 for 30 sec. the flow-through was discarded, then it Centrifuged at 14000rpm for 3 min again, finally DNA was eluted using dH2O (100 µl). Healthy DNA was extracted from whole blood using (Genaid extraction kit), in briefly ; A 300 µl of frozen blood was transferred to eppendorf tube, then 40 µl of proteinase k was added and incubated it at 60 C for 20 min, the GB buffer was added ( 200 µl ) and it shaking vigorously, after this absolute ethanol was added (200 µl) with mixed it by shaking, then it centrifuged at 15000 rpm for 5 mints. the Supernatant was transferred to GD column, and centrifuged at 15000 rpm for 1 min. the flow -rate was discarded and 400 µl of W1 buffer was added, then centrifuged at 15000 rpm for 1 min, the Fallow rate was discarded also, about 600 µl of wash buffer was added, then centrifuged at 15000 rpm for 1 min. columns were Re-centrifuged after discarded flow ate for 5 min at the same speed to dry column, finally 100 µl of d H2O was added to column and left 2 min at room temperature to absorb it. DNA eluted in new eppendorf tube by centerfield column for 2 min at15000 rpm.
After DNA extraction; consternation and purity of DNA were estimated using nanodrpe. PCR conditions were performed as a following; primers that used F- GGGTCTTTGCACCATCTCTG , R- TCCAGTCTTCCAAGCTCTTCA. PCR annealing temperature was 57C for 20 sec.
SSCP technique, PCR products were denaturation using SSCP dye (EDTA, formamid and bromophynol blue) 1/1 V:V in water bath for 5 min at 95C then its child in ice for 2 min.;
SSCP electrophoresis, the products were electrophoresis as a following About 10 µl of the samples into wells of an 8% acrylamide/bis gel (37.5:1), containing 7% glycerol, and 1x TBE buffer. And for recipe a 20 x 20 x 0.1 cm gel format. 8 ml of 40% acrylamide/bis (37.5:1) mixed with 8 ml of 5x TBE , 2,8 ml 100% glycerol, then 40 µl TEMED and 400 µl of 10% ammonium per sulfate were added with 20.8 ml of dH2O After gel was casting sample were loaded and Run under the following conditions.
Buffer 1x TBE, Buffer temperature 10°C ,Run time 3.5 hours and 100V. Then gel was staining using ethedium bromide for 15 min.
The results of present study show that the PCR product of micro-RNA gene was 197 pb as show in figure (1-A) all patients and control samples give positive amplification , this mean that no deletion nor duplication in our population study, for more investigation pcr products were analysis using SSCP technique to detection haplotype polymorphism in this gene, the results show that there were a clear results in haplotype (d) which appeared in patients (100)% and disappeared in control. Also haplotype A was appeared in all control but in patients it was appeared in 75% only as show in table (1) and figure (1,B,C).
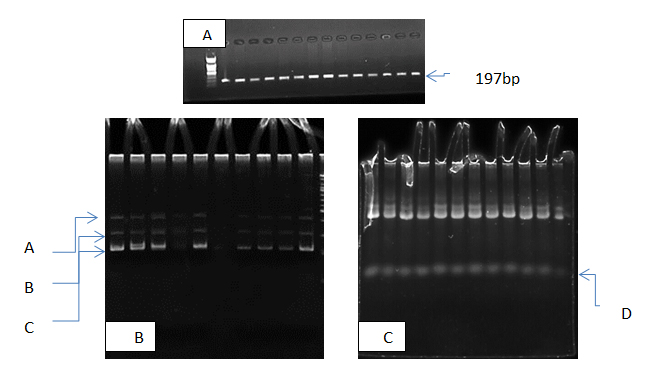
Fig. 1. Electrophoresis pattern of micrRNA-146a gene in study subjects ; A PCR products 197bp., B SSCP pattern of control samples, C SSCP pattern of patients samples
Table (1):
The Haplotypes distribution and odd ratio of MIR-146a gene polymorphism for patients and control.
Haplotypes name |
Patients% |
Control% |
OD ratio |
CI 95% |
P-value |
|---|---|---|---|---|---|
A |
75 |
100 |
67.8874 |
4.0677 -1133.0016 |
0.0012* |
B |
100 |
100 |
1 |
0.0196 – 50.8937 |
1 |
C |
100 |
100 |
1 |
0.0196 – 50.8937 |
1 |
D |
100 |
0 |
40401 |
793.83 – 2056156.37 |
<0.0001* |
*significant at p value <0.05
These differences in haplotype D may be because SNPs in this DNA sequences, or inverse DNA segments or changes in chemical structure of DNA component that lead to mutations, as show in previous studies micro-RNA was a short sequence of RNA and any changes in its genes may be effect in it and in its functions, these variation deal with some studies improved that the SNPs within miRNA sequences, or its pathway genes, and miRNA binding sites that have been found to be correlated with cancer susceptibility like Jazdzewski et al., (2008) how improved associated micro-RNA146a SNPs with tumorigenesis through somatic mutation. Also some SNPs such as hsa-miR-146 rs2910164 and hsa-miR-196a2 rs11614913 association with breast cancer patients in north Indian women (Bodal et al., 2017). the association between miRNA- SNPs and cancer risk has been reported predominantly by Horikawa et al. (2007) whom evaluated SNPs in miRNA-related genes and the risk of bladder cancer and renal cell carcinoma. They found that two SNPs in the GEMIN4gene are associated with altered renal cell carcinoma risk. Also Song and Chen (2011) reported some SNPs associated with cancer risk, one of these was rs895819 in miRo-27a gene which related with breast cancer.
In Iraq this is the first study deal with micro-RNA genes polymorphisms in breast cancer thus we need more investigations about the role of micro-RNA genes and its biogenesis pathway in cancer risk in Iraqi population however . there are many factors effected in DNA sequences like environment, lifestyle, nutrition, stress , and different physical and chemicals factors these factors may be led to these SNPs, the review of literature show that these factors have been effected in some genes related with some disease in Iraq, other studies deal with the same breast cancer samples which used in present study show that there were more than one genes related to breast cance;r thus the present study was conducted to clarify regulatory gene mico-RNA146a genotype in breast cancer tissue, the present results show significant differences in tow haplotype from total 5 type, this can be explanation that breast cancer in Iraq is multifactorial disease that synergist to causes this disease. The variation in Iraqi population may be because the life style of individuals or elevated oxidative stress which causes accumulation of ROS, this causes different mutations in DNA sequence, gene expression and its regulation, also studies which deal with Iraqi population improve that Iraqi patients suffered from contributed more than one factor which causes disease (Al-Terehi et al.,2015). The environmental pollution have been main role in incidence disease in Iraqi patients like heavy metals (Al-Terehi et al.,2015).
- Akbani, R., Ng, P.K.S., Werner, H.M.J., Shahmoradgoli, M., Zhang, F., Ju, Z.,Liu, W., Yang, J.-Y., Yoshihara, K., Li, J., et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun., 2014; 5: 3887.
- Han, L., Yuan, Y., Zheng, S., Yang, Y., Li, J., Edgerton, M.E., Diao, L., Xu, Y., Verhaak, R.G.W., and Liang, H. The Pan-Cancer analysis of pseudogene expression reveals biologically and clinically relevant tumour subtypes. Nat. Commun. 2014; 5: 3963.
- Hoadley, K.A., Yau, C., Wolf, D.M., Cherniack, A.D., Tamborero, D., Ng, S., Leiserson, M.D.M., Niu, B., McLellan, M.D., Uzunangelov, V., et al.; Cancer Genome Atlas Research Network. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell, 2014; 158: 929–944.
- International Human Genome Sequencing Consortium: Finishing the euchromatic sequence of the human genome. Nature. 2004; 431:931-45.
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494-8.
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008; 9: 219-30.
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002; 99: 15524-9.
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic micro- RNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005; 65: 9628-32.
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006; 103: 2257-61.
- Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008; 378: 492-504.
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005; 120: 635-47.
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007; 21: 1025-30.
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong Y, et al. let-7 regulates self-renewal and tumorigenicity of breast cancer cells. Cell. 2007; 131: 1109-23.
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon- Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009; 23: 2839-49.
- Chen C, Ridzon D, Lee CT, Blake J, Sun Y, Strauss WM. Defining embryonic stem cell identity using differentiation- related microRNAs and their potential targets. Mamm Genome. 2007; 18: 316-27.
- Al-Terehi1, M. al-kilabi2, L.H., AL –Mamoori1, A., Al-Jboori, M.J. , Al-Saadi1, A H. Zaidan H.K. Some Heavy Metals Concentrations in Tumor Tissue, International Journal of ChemTech Research CODEN (USA): IJCRGG 2016, 9(3): 407-411.
- Al-Terehi, Haider K. Zaidan2, Ayad M.J. AL –Mamoori2; Ali Hmood Al-Saadi2, Israa Harjan Effective of different factors on trace elements concentrations in Iraqi lactating mother’ smilk nternational Journal of Pharm Tech Research, 2015; 8(10): pp 151-157.
- Al-Terehi M., Ghaleb R, Al-Oubaidy SH., Al-Saadi A.,. Zaidan H. Study TNF-± gene polymorphism in Type 1 Diabetic Patients Using ARMS technique Journal of Chemical and Pharmaceutical Science CPS, 2016; 9(3); 1107-1111.
- Al-Terehi M., Hasan A., AL-Jboory J M. Al-Saadi1 A., Zaidan H., obiad S. Haplotype Polymorphisms in Cytokines Genes Using Pcr-Sscp Technique in Iraqi Breast Cancer Patients Der Pharma Chemica, 2016, 8(22): 27-31.
- Jazdzewski K, Murray EL, Franssila K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma [J] Proc Natl Acad Sci USA. 2008;105(20):7269–7274. [PMC free article] [PubMed]
- Horikawa Y, Wood CG, Yang H, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma [J]Clin Cancer Res. 2008; 14(23):7956–7962.[PMC free article] [PubMed].
- Bodal, V.K. Sangwan S, Bal M.S. , Kaur M., Sharma S., and Kaur B. Association between Microrna 146a and Microrna 196a2 Genes Polymorphism and Breast Cancer Risk in North Indian Women Asian Pac J Cancer Prev. 2017; 18(9): 2345–2348.
- Song F-J, Chen K-X. Single-nucleotide polymorphisms among microRNA: big effects on cancer. Chinese Journal of Cancer. 2011; 30(6):381-391. doi:10.5732/cjc.011.10142.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


