ISSN: 0973-7510
E-ISSN: 2581-690X
Antimicrobial resistance (AMR) is the capability of a microorganism to neutralize the harmful effects of a drug. AMR is an increasing health problem worldwide. UTIs are among the most common infection in human accounting for 150 million cases globally. E. coli is the most common pathogen responsible for these infections. The uropathogens are getting resistant to commonly used antibiotics. The current study was designed to evaluate the antibiotic resistance pattern of the uropathogens against commonly administered antibiotics in patients visiting Rafha central Hospital, Rafha city, Saudi Arabia. The study was done retrospectively and the data was collected from the hospital lab from January 2016 to December 2017. During that period, 623 positive cases were observed. E. coli was the most prevalent UTI pathogen. Resistance against 27 commonly used antibiotics was studied. Among β-Lactam antibiotics, increasing resistance was observed except for Augmentin. The imipenem was relatively more effective. Among non-β-Lactam group, least resistance was seen against Vancomycin and Amikacin. Overall increase in antibiotic resistance was observed in the current study with some exceptions. It is therefore recommended that the routine urine cultures must be done and the resistance pattern in the region must regularly be monitored.
Antimicrobial resistance (AMR), Uropathogens, Pattern, Urinary Tract Infection (UTI), Escherichia coli, Prevalence, Susceptible
Antimicrobial resistance (AMR) is the capability of a microorganism to neutralize the harmful effects of a drug which is used to stop their growth or kill them1. AMR is considered as one of the utmost universal threats to human health. Microorganisms that are resistant to one or more drugs are harder to treat, necessitating the use of alternative drug or higher doses of the same drug, which can be expensive or even more toxic.
Urinary tract infections (UTI) are the most common and serious health problem among, both outpatients and hospitalized patients affecting millions of individuals worldwide2. Because the urinary tract is in direct contact with the outer environment, it is more likely to get infected3. About 150 million cases of UTI are estimated every year worldwide4. The disease affects all age groups with manifestations varying from symptomatic cystitis to pyelonephritis and septicemia2. Improperly treated UTI can result in substantial morbidity and mortality5.
Women usually have more incidences than men due to the anatomical organization of their genitourinary tract6. It has been estimated that at least 30% of all women get a UTI at some point during their lifetime7. The other UTI causing factors consist of long time catheterization, poor hygiene, sexual inter course during menstruation period and pregnancy8.
The most common pathogen causing UTI is Escherichia coli9 followed by Klebsiella pneumonie, Staphylococcus aureus and Pseudomonas aeruginosa10. During laboratory investigation, a bacterial infection of the urinary tract is considered positive if it shows bacterial cells greater than 10 5 per milliliter of urine.
The UTIs treatment depends on the age and sex of the patient, and the causative agent. It also depends on the site of infection i.e. lower or upper urinary tract infection. Cotrimoxazole (Trimethoprim/sulphamethoxazole) and ciprofloxacin are the most commonly used drugs for the treatment of UTI. The other commonly used drugs include fluoroquinolones, β-lactams (occasionally with β-lactamase inhibitors), cephalosporin and nitrofurantoin11. Recent studies show that resistance to many antimicrobials including the fluoroquinolones is increasing12. The increase in bacterial resistance to fluoroquinolone is multifactorial. With the increasing trend of antibiotic-resistance in E. coli, the management of urinary tract infections is likely to become complicated with limited therapeutic options.
This retrospective study was done to assess the current antibiotic resistance pattern among the UTI pathogens against commonly prescribed antibiotics in patients visiting Rafha central Hospital, Rafha city, Saudi Arabia.
The study was done retrospectively on the antibiotic resistance pattern of uropathogens for 2 year (January 2016 to December 2017). The required culture and sensitivity data was collected from the records of Microbiology laboratory of Rafha Central Hospital, Rafha, Saudi Arabia. Approval from the Institutional Ethics Committee was obtained prior to the study.
A total of 2204 urine samples during the two years were brought to the lab for culture sensitivity testing. The samples were collected, cultured and antibiotic susceptibility was determined according to the Standard Clinical Laboratory procedures of the Lab. Briefly, urine samples were collected in specified sterile containers and brought to lab. Each sample was cultured on Blood Agar medium and MacConkey Agar medium using the calibrated loop technique. The plates were incubated overnight at 37°C. Bacterial growth > 105 cfu/ml was considered as significant. For identification of the bacterial isolates, conventional methods were used. The antimicrobial sensitivity testing was done using the standard Kirby-Bauer disc diffusion technique on Mueller Hinton agar medium according to the CLSI guidelines. The antibiotics discs used for the AST were from MASTRING-STM, Mast Diagnostics, UK and included: Penicillin G (10 units), Ampicillin (10 µg), Augmentin (30 µg), Oxacillin (1 µg), Piperacillin (100 µg), Cephalothin (30 µg), Cefoxitin (30 µg), Cefuroxime (30 µg), Ceftazidime (30 µg), Ceftriaxone (30 µg), Cefotaxime (30 µg), Cefepime (30 µg), Aztreonam (30 µg), Imipenem (10 µg), Tetracycline (30 µg), Amikacin (30 µg), Gentamicin (10 µg), Neomycin (30 µg), Erythromycin (15 µg), Chloramphenicol (30 µg), Nalidixic acid (30 µg), Cip/Norfloxacin (10 µg), Nitrofurantoin (300 µg), Cotrimoxazole (50 µg), Vancomycin (30 µg), Polymyxin B (300 units), Fusidic acid (10 µg)
The results were calculated as frequency and percentage. Chi square test was used to find any significant correlation between different factors. The one tailed p values were calculated online at http://vassarstats.net/tab2x2.html
PCR
PCR was done to identify microbial strains and detect few resistant genes in some of the clinical isolates in order to compare our molecular methods with the standard microbial methods performed in hospital laboratory. For that purpose, ten samples of each of E. coli, Klebsiella, Pseudomonas and Coagulase negative Staphylococcus species were analyzed.
DNA Extraction
A single medium sized colony was suspended in 50 microliters of low TE buffer in a 200 microliter PCR tube and boiled at 95 °C for 5 minutes. It was then cooled and centrifuged at 5000 rpm for 1 minute and the supernatant containing DNA was transferred to a new tube. One microliter of this DNA was used in 20 microliter PCR mixture. The PCR mixture contained 1X PCR buffer (with KCl), 1.5mM MgCl2, 0.2mM dNTPs, 0.5mM each primer (Table 1), 0.5 units of Taq polymerase (Thermo Scientific UK). Cycling conditions for PCR were: Initial denaturation at 95°C for 5 minutes and then 35 cycles of 95°C for 15 seconds, annealing (Table 1) for 15 seconds and 72°C for 30 seconds, followed by a final extension at 72°C for 5 minutes. Five microliters of PCR product were loaded on a 2% agarose gel in 1X TBE buffer to confirm the presence of PCR product.
Table (1):
Oligonucleotide primers used
| Primer Name | Primer Sequence | Target | Product Size | Annealing temp |
|---|---|---|---|---|
| M12-F | 5′- GTGATCTCCAGCTACCGCTA-3′ | E. coli | 200 | 55° C |
| M12-R | 5′- CGTTGCAAACTGACGCTCTT-3′ | |||
| PsA-F | 5’-TTCCGGTGAAGGTGCCAATG-3′ | Pseudomonas Aeruginosa | 297 | 57° C |
| PsA-R | 5’-AGGTAGCGCTGAACGGCCTT-3′ | |||
| KN-F | 5’-GTCATGCTCTCGGTGCTGTT-3′ | Klebsiella pneumoniae | 236 | 55° C |
| KN-R | 5’-GACACCGCGGTCATCATTAC-3′ | |||
| CNS-F | 5’-TATCCACGAAACTTCTAAAACAACTGTTACT-3′ | Coagulase Negative Staphylococcus | 204 | 57° C |
| CNS-R | 5’-TCTTTAGATAATACGTATACTTCAGCTTTGAATTT-3′ | |||
| 16S-F | 5′-CTAGTAATCGCGGATCAGCAT -3′ | 16s RNA | 174 | 54° C |
| 16S-R | 5′- GATACGGCTACCTTGTTACGACTT-3′ | |||
| SUL1-F | 5’-TTCGGCATTCTGAATCTCAC-3′ | Sulphonamide resistant gene | 822 | 54° C |
| SUL1-R | 5’-ATGATCTAACCCTCGGTCTC-3′ | |||
| ere(A)-F | 5’-GCCGGTGCTCATGAACTTGAG-3′ | Erythromycin resistant gene | 419 | 57° C |
| ere(A)-R | 5’-CGACTCTATTCGATCAGAGGC-3′ | |||
| tetA-F | 5’-GGTTCACTCGAACGACGTCA-3′ | Tetracycline resistant gene | 577 | 55° C |
| tetA-R | 5’-CTGTCCGACAAGTTGCATGA-3′ | |||
| DfrA1-F | 5’-GGAGTGCCAAAGGTGAACAGC-3′ | Trimethoprim resistant gene | 367 | 57° C |
| DfrA1-R | 5’-GAGGCGAAGTCTTGGGTAAAAAC-3′ | |||
| CatA1-F | 5’-AGTTGCTCAATGTACCTATAACC-3′ | Chloramphenicol resistant gene | 547 | 54° C |
During the two years study period, 2204 urine samples (1,028 in 2016 and 1,176 in 2017) were brought and processed in the lab Out of which, 623 (28.27%) samples showed significant growth of pathogen. Remaining 1,581 samples were either sterile or had a very low bacterial count.
E. coli was the most prevalent UTI pathogen (43.3%) isolated during the study period followed by Klebsiella (15.9%) and Staphylococci (15.2%); Citrobacter was the least prevalent (1.1%). The prevalence of E. coli and Acinetobacter species isolated in 2016 were significantly higher than those of 2017 while on the other hand, the prevalence of Enterococci, Klebsiella and Pseudomonas species were higher during 2017 (Table 2).
Table (2):
Frequency and percentage of different bacterial strains isolated during the two years
Total N (%) |
2016 N (%) |
2017 N (%) |
p value |
|
|---|---|---|---|---|
Acinetobacter |
20 (3.2) |
19 (7.1) |
1 (0.3) |
<0.0000 |
Citrobacter |
7 (1.1) |
5 (1.9) |
2 (0.6) |
— |
E. coli |
270 (43.3) |
130 (48.5) |
140 (39.4) |
0.0146 |
Enterobacter |
10 (1.6) |
3 (1.1) |
7 (2) |
— |
Enterococci |
55 (8.8) |
14 (5.2) |
41 (11.5) |
0.0038 |
Klebsiella |
99 (15.9) |
33 (12.3) |
66 (18.6) |
0.0213 |
Proteus |
25 (4) |
9 (3.4) |
16 (4.5) |
— |
Pseudomonas |
42 (6.7) |
10 (3.7) |
32 (9) |
0.0062 |
Staphylococci |
95 (15.2) |
45 (16.8) |
50 (14.1) |
— |
Total |
623 |
268 |
355 |
In our lab E. coli, Klebsiella, Pseudomonas and Coagulase negative Staphylococcus species were detected by PCR amplification and on agarose gel electrophoresis along with five genes responsible for resistance to five common antibiotics (Fig. 1).
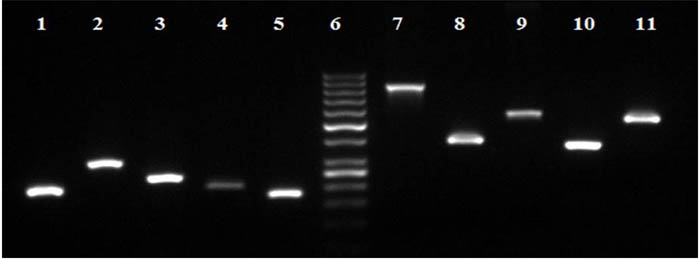
Fig. 1. Agarose gel electrophoresis showing PCR bands for the identification of some of the pathogens and antimicrobial resistant genes. Lanes 1, E. Coli (200bp); Lane 2, Pseudomonas Aeruginosa (297bp); Lane 3, klebsiella pneumoniae (236bp); Lane 4, Coagulase Negative staphylococcus (204bp); Lane 5, 16S RNA (174bp), Lane 6, GeneRuler 50bp DNA ladder (Thermo Fisher Scientific); Lane 7, Sulphonamides (822bp); Lane 8, Erythromycin (419bp); Lane 9, Tetracycline (577bp); Lane 10, Trimethoprim (367bp); and Lane 11, Chloramphenicol (547bp)
In the microbiology laboratory of the Rafha Central Hospital, a total of 27 antibiotics were used to study the antibiotic susceptibility patterns out of which 14 belonged to β-Lactam group (Table 3). The average resistance against penicillin group was 73.4% during 2016 and 78.4% during 2017. The resistance increased significantly against penicillin G (p = 0.0128) and oxacillin (p = 0.00507) while decreased against Augmentin (p = 0.000011) during 2017. Among Cephalosporins, the resistance ranged from 35% against Cefoxitin to 76% against Cephalothin. A significant increase in resistance was seen against Cefoxitin (p = 0.0036), cefuroxime (p = 0.0411), Ceftazidime (p = 0.0221) and Cefotaxime (p = 0.00001) during 2017. The monobactam Aztreonam also showed similar results (p = 0.00034).
Table 3. AMR pattern against b-Lactam antibiotics
| Antibiotic class | Penicillin group | Cephalosporins | ||||||||||||
| 1st generation Cephalothin | 2nd generation Cefoxitin | 2nd generation Cefuroxime | 3rd generation Ceftazidime | 3rd generation Ceftriaxone |
3rd generation Cefotaxime |
4th generation Cefepime |
Monobactam Aztreonam |
Carbapenem Imipenem |
||||||
| Agent | Penicillin G | Ampicillin | Augmentin | Oxacillin | Piperacillin | |||||||||
| 2016 | ||||||||||||||
| Resistant | 17 | 179 | 125 | 30 | 143 | 167 | 57 | 8 | 52 | 26 | 25 | 45 | 59 | 18 |
| Susceptible | 5 | 49 | 44 | 14 | 63 | 54 | 108 | 9 | 71 | 21 | 23 | 73 | 73 | 190 |
| Total | 22 | 228 | 169 | 44 | 206 | 221 | 165 | 17 | 123 | 47 | 48 | 118 | 132 | 208 |
| %age | 77% | 79% | 74% | 68% | 69% | 76% | 35% | 47% | 42% | 55% | 52% | 38% | 45% | 9% |
| 2017 | ||||||||||||||
| Resistant | 174 | 257 | 131 | 55 | 158 | 212 | 114 | 84 | 112 | 29 | 35 | 14 | 135 | 22 |
| Susceptible | 10 | 52 | 116 | 6 | 63 | 91 | 121 | 33 | 94 | 10 | 1 | 29 | 76 | 174 |
| Total | 184 | 309 | 247 | 61 | 221 | 303 | 235 | 117 | 206 | 39 | 36 | 43 | 211 | 196 |
| %age | 95% | 83% | 53% | 90% | 71% | 70% | 49% | 72% | 54% | 74% | 97% | 33% | 64% | 11% |
| p value | 0.0128 | 0.105 | 0.00001 | 0.00507 | 0.35789 | 0.0937 | 0.0036 | 0.0411 | 0.0221 | 0.0535 | 0.000001 | 0.3233 | 0.00034 | 0.2425 |
Among non-β-Lactam group, least resistance was seen against Amikacin (23.8%) and vancomycin (26.5%) during the two years whereas Fusidic acid and Erythromycin faced maximum resistance (92% each). Gentamicin showed decreased (p = 0.026) while chloramphenicol and Ciprofloxacin /Norfloxacin showed increased resistance (p = 0.0009 and p = 0.006 respectively) during 2017 (Table 4).
Table (4):
AMR pattern against non-β Lactam antibiotics
Protein synthesis inhibitors
| Antibiotic class | Tetracycline | Aminoglycosides | Macrolides | Chloramphenicol | Quinolone | Nitrofurantoin | Cotrimoxazole | Glycopeptide | Polypeptide | Steroids | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Agent | TC | Amikacin | Gentamicin | Neomycin | Erythromycin | Nalidixic acid | Cip/Norfloxacin | Vancomycin | Polymyxin B | Fusidic acid | |||
| 2016 | |||||||||||||
| Resistant | 44 | 50 | 92 | 8 | 24 | 15 | 171 | 123 | 81 | 173 | 9 | 6 | 26 |
| Susceptible | 12 | 158 | 119 | 1 | 2 | 22 | 51 | 116 | 133 | 59 | 38 | 3 | 3 |
| Total | 56 | 208 | 211 | 9 | 26 | 37 | 222 | 239 | 214 | 232 | 47 | 9 | 29 |
| %age | 79% | 24% | 44% | 89% | 92% | 41% | 77% | 51% | 39% | 75% | 19% | 67% | 90% |
| 2017 | |||||||||||||
| Resistant | 18 | 44 | 88 | 29 | 81 | 71 | 219 | 174 | 113 | 235 | 18 | 26 | 21 |
| Susceptible | 7 | 143 | 168 | 14 | 7 | 28 | 52 | 103 | 155 | 79 | 37 | 15 | 1 |
| Total | 25 | 187 | 256 | 43 | 88 | 99 | 271 | 277 | 268 | 314 | 55 | 41 | 22 |
| %age | 72% | 24% | 34% | 67% | 92% | 72% | 81% | 63% | 42% | 75% | 33% | 63% | 95% |
| P value | 0.3531 | 0.5003 | 0.0260 | 0.1912 | 0.6643 | 0.0009 | 0.1794 | 0.0060 | 0.1933 | 0.5099 | 0.0920 | 0.5874 | 0.4167 |
The AMR pattern of tested antibiotics against various isolated strains
E Coli
A high percentage of isolated E. coli strains were resistance to penicillin group during the study period with average resistance of 78%. The resistance against Augmentin decreased significantly in 2017 (p = 0.0011, Table 5). Similar results were also seen against Cephalothin among cephalosporin group (p = <0.0001, Table 5). Cefoxitin (second generation drug) faced the lowest resistance among cephalosporin during 2016 which increased slightly during 2017 but was statistically insignificant. The Cefepime (4th generation) showed relatively better results than most other Cephalosporins. The overall resistance against cephalosporin was >50%.
Table (5):
The AMR pattern among different isolated bacteria
| E. coli | Klebsiella sp. | Enterococci | Staphylococcus | Psudomonas | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | p | 2016 | 2017 | p | 2016 | 2017 | p | 2016 | 2017 | P | 2016 | 2017 | p | |
| Res/Sus (Res %) | Res/Sus (Res %) | Res/Sus (Res %) | Res/Sus (Res %) | Res/Sus (Res %) | Res/Sus (Res %) | Res/Sus (Res %) | Res/Sus (Res %) | Res/Sus (Res %) | Res/Sus (Res %) | ||||||
| Penicillin G | —– | 53/0 (100) | —– | —– | 29/0 (100) | —– | 5/3 (63) | 21/3 (88) | —– | 6/2 (75) | 35/3 (92) | —– | —– | 21/0 (100) | —– |
| Ampicillin | 88/15 (85) | 104/18 (85) | —– | 23/2 (92) | 52/5 (91) | —– | 7/7 (50) | 16/15 (52) | —– | 18/18 (50) | 33/11 (75) | 0.0186 | 8/0 (100) | 28/0 (100) | —– |
| Augmentin | 68/21 (76) | 47/41 (53) | 0.0011 | 15/3 (83) | 31/17 (65) | —– | 4/1 (80) | 6/22 (21) | 0.0214 | 4/5 (44) | 14/22 (39) | —– | 8/1 (89) | 25/3 (89) | —– |
| Oxacillin | —– | —– | —– | —— | —– | —– | 4/2 (67) | 16/4 (80) | —– | 21/12 (64) | 26/1 (96) | 0.002 | —– | 3/0 (100) | —– |
| Piperacillin | 89/22 (80) | 72/25 (74) | —– | 19/11 (63) | 35/10 (78) | —– | 1/0 (100) | 6/5 (55) | —– | 3/8 (27) | 9/7 (56) | —– | 3/7 (30) | 21/8 (72) | 0.0235 |
| Cephalothin | 94/9 (91) | 82/36 (69) | 0.00003 | 18/6 (75) | 46/11 (81) | —– | 4/1 (80) | 16/20 (44) | —– | 11/28 (28) | 22/21 (51) | 0.0287 | 9/0 (100) | 23/0 (100) | —– |
| Cefoxitin | 14/78 (15) | 19/60 (24) | —– | 6/10 (38) | 26/21 (55) | —– | 2/1 (67) | 16/11 (59) | —– | 6/3 (67) | 24/11 (69) | —– | 8/1 (89) | 21/4 (84) | —– |
| Cefuroxime (CXM) | —– | 23/21 (52) | —– | —– | —– | 7/5 (58) | —– | —– | 12/3 (80) | —– | 0/1 (0) | 19/0 (100) | 0.0499 | ||
| Ceftazidime (CAZ) | 27/48 (36) | 26/54 (33) | —– | 5/11 (31) | 19/17 (53) | —– | 1/1 (50) | 14/6 (70) | —– | —– | 28/1 (97) | —– | 1/7 (13) | 12/10 (55) | 0.0473 |
| Ceftriaxone (CRO) | —– | 6/8 (43) | —– | —– | —– | —– | 6/1 (86) | 2/1 (67) | —– | 7/13 (35) | —– | —– | —– | 10/0 (100) | —– |
| Cefotaxime (CTX) | —– | 6/0 (100) | —– | —– | —– | —– | 4/2 (67) | 5/1 (83) | —– | 6/15 (29) | 7/0 (100) | 0.0014 | —– | 8/0 (100) | —– |
| Cefepime (CPM) | 7/28 (20) | 4/11 (27) | —– | 7/9 (44) | 0/4 (0) | —– | 5/2 (71) | 3/7 (30) | —– | 12/22 (35) | 4/6 (40) | —– | 0/1 (0) | 1/1 (50) | —– |
| Aztreonam | 33/46 (42) | 51/36 (59) | 0.0219 | 6/11 (35) | 31/15 (67) | 0.0226 | 1/0 (100) | 7/3 (70) | —– | 6/2 (75) | 16/1 (94) | —– | 2/6 (25) | 17/10 (63) | —– |
| Imipenem | 3/112 (3) | 8/77 (9) | 0.0385 | 1/30 (3) | 6/33 (15) | —– | —– | 1/12 (8) | —– | 2/8 (20) | 3/17 (15) | —– | 0/10 (0) | 2/18 (10) | —– |
| Tetracycline | 22/4 (85) | 7/0 (100) | —– | 5/1 (83) | 2/1 (67) | —– | —– | —– | —– | —– | 7/3 (70) | —– | 0/3 (0) | —– | —– |
| Amikacin | 11/100 (10) | 11/75 (13) | —– | 7/24 (23) | 6/31 (16) | —– | 1/0 (100) | 3/5 (38) | —– | 2/10 (17) | 7/9 (44) | —– | 1/9 (10) | 6/15 (29) | —– |
| Gentamicin | 26/69 (27) | 21/73 (22) | —– | 11/13 (46) | 13/36 (27) | —– | 8/1 (89) | 9/21 (30) | 0.0026 | 14/21 (40) | 22/13 (63) | 0.0467 | 4/4 (50) | 12/14 (46) | —– |
| Neomycin | —– | —– | —– | —– | 7/3 (70) | —– | —– | —– | —– | —– | 1/4 (20) | —– | 1/1 (50) | 10/2 (83) | —– |
| Tobramycin | —– | 5/10 (33) | —– | —– | 3/3 (50) | —– | —– | —– | —– | —– | 7/2 (78) | —– | —– | 2/3 (40) | —– |
| Erythromycin | —– | 16/0 (100) | —– | —– | 13/0 (100) | —– | 1/0 (100) | 15/4 (79) | —– | 17/0 (100) | 25/3 (89) | —– | —– | 8/0 (100) | —– |
| Chloramphenicol | —— | 24/8 (75) | —– | —– | 12/1 (92) | —– | —– | 6/9 (40) | —– | 6/21 (22) | 8/9 (47) | —– | —– | 15/3 (83) | —– |
| Nalidixic acid | 67/33 (67) | 81/26 (76) | —– | 15/13 (54) | 37/16 (70) | —– | 12/1 (92) | 27/2 (93) | —– | 40/1 (98) | 27/2 (93) | —– | 6/0 (100) | 27/1 (96) | —– |
| Cip/Norfloxacin | 50/61 (45) | 64/44 (59) | 0.0243 | 10/19 (34) | 28/27 (51) | —– | 10/3 (77) | 23/6 (79) | —– | 24/15 (62) | 24/8 (75) | —– | 1/8 (11) | 17/13 (57) | 0.0187 |
| Nitrofurantoin | 18/77 (19) | 29/83 (26) | —– | 15/14 (52) | 25/21 (54) | —– | 4/10 (29) | 9/17 (35) | —– | —– | —– | —– | 5/1 (83) | 25/2 (93) | —– |
| Cotrimoxazole | 73/31 (71) | 88/35 (72) | —– | 19/7 (73) | 44/16 (73) | —– | 10/1 (91) | 26/12 (68) | —– | 25/14 (64) | 34/11 (76) | —– | 9/0 (100) | 26/0 (100) | —– |
| Clindamycin | —– | 11/0 (100) | —– | —– | 8/0 (100) | —– | —– | 17/4 (81) | —– | —– | 24/4 (86) | —– | —– | 5/1 (83) | —– |
| Vancomycin | —– | —– | —– | —– | 1/2 (33) | —– | 1/11 (8) | 3/16 (16) | —– | 4/26 (13) | 10/15 (40) | 0.0252 | —– | 3/0 (100) | —– |
Res; Resistant, Sus; susceptible
Imipenem was the drug of choice followed by Amikacin and Nitrofurantoin during the study period as they were effective against most of the isolated E coli strains. Maximum (100%) resistance was seen against Tetracycline, erythromycin and clindamycin. Resistance against imipenem, Aztreonam and Cip/Norfloxacin increased significantly during the study period.
Klebsiella spp.
Penicillin group was poor against isolated klebsiella species facing 84% resistance overall with penicillin G being 100 ineffective. Cephalosporins were the same with more than 50% resistance except Cefoxitin, ceftazidime and Cefepime (38%, 31% and 44% respectively) during 2016. Imipenem was much better with average 9% resistance followed by Amikacin with 19.5% average resistance. Erythromycin and clindamycin faced 100% resistance followed by chloramphenicol (92%) and Cotrimoxazole (73%)
Enterococci
Penicillins were less effective here too, facing 66% resistance overall. Resistance against Augmentin decreased significantly in 2017 from 80% to 21% (p = 0.0214). Cephalosporins also had poor efficacy facing 64% resistance. Among non-beta lactam group, imipenem was the most effective with 8% resistance only. The resistance against gentamicin decreased significantly in 2017 (p = 0.0026, Table 5)
Staphylococci
A higher proportion of staphylococcus species were resistant against Penicillins during the study period with average resistance of 63%; piperacillin faced least resistance (27%) in 2016 while oxacillin was almost ineffective with 96% resistance in 2017. The resistance increased significantly against Ampicillin and oxacillin during the study period (p = 0.0186 and p=0.002 respectively, Table 5).
Staphylococci expressed a variety of resistance against Cephalosporins (average resistance 57%) from 28% against Cephalothin in 2016 to 100% against cefotaxime in 2017. Resistance against all the Cephalosporins used in the study increased during the two years but those Cephalothin and Cefotaxime were statistically significant (p=0.0287 and p=0.0014 respectively). Imipenem again was the most effective antibiotic among the non-β-lactams with 16.7% resistance followed by vancomycin (25.5), Amikacin and chloramphenicol (32% each). Nalidixic acid was the least effective against this group of pathogens facing average resistance of 95.7% followed by erythromycin (93%) and Aztreonam (88%, Table 5).
Pseudomonas
Most penicillin like penicillin G, ampicillin and oxacillin were totally useless against these pathogens while Augmentin faced 89% resistance. Only piperacillin was a little better facing 30% resistance which increased to 72% during 2017 which proved to be a significant change (p=0.0235). Cephalosporins also proved to be almost ineffective with significant increase in resistance against cefuroxime (p=0.0499) and ceftazidime (p=0.0473) during the study period. Imipenem was the most effective drug against Pseudomonas with 6.6% overall resistance followed by amikacin (22.6% resistance). Most other non-β-lactams faced 80-100% resistance (Table 5).
This is first study to appraise the antimicrobial resistance pattern among bacterial pathogens isolated from patients with urinary tract infections in Rafha, Kingdom of Saudi Arabia.
After upper respiratory tract infections, UTI are the most common infections worldwide 13, 14, 15, which are therefore, important cause of morbidity and mortality and cost over 6 billion US dollars annually worldwide13. For that reason, the uropathogens and their AMR pattern must be studied to decide effective treatment of the infection14.
It is a well-known fact that the antibiotic resistance in community acquired pathogens is an ever increasing phenomenon16, 17, 18. Increasing rates of antibiotic resistance among most of the pathogenic bacteria, including Gram negative bacteria, decrease the options for the treatment of deadly infections. The widespread antibiotic resistant pathogenic bacteria are now a serious public health concern worldwide16. Infections caused by multidrug resistant bacteria can result in longer hospital stays and increased mortality 19, 20, 21. The resistance was found to be highly prevalent during the current study. Many drugs showed increased resistance in 2017 as compared to that seen in 2016. These drugs include Penicillin G, Oxacillin, Cefoxitin, Cefuroxime, Ceftazidime, Cefotaxime, Aztreonam, Chloramphenicol and Ciprofloxacin/Norfloxacin. This is an alarming sign for the concerned authorities of the local health department and suggests changes in treatment options. Augmentin and gentamicin however showed an opposite trend and were relatively more effective during 2017 than 2016. Imipenem, a member of carbapenem group was found to be the most effective antibiotic especially among the gram negative organisms with resistance not exceeding 15%. El-Kersh et al., 2015 has reported similar results22.
E. coli has been reported to be the most common uropathogens worldwide ranging from 36% to 77%22, 23. During the study period, E. coli was the most common UTI pathogen (43.3%) followed by Klebsiella (15.9%) and Staphylococci (15.2%). Among the resistant E. coli, 22% were ESBL producers in 2016 and 28% in 2017.
All the gram negative bacteria (E. coli, Klebsiella and Pseudomonas) isolated in the current study were 100% resistant to penicillin G. The E. coli, Klebsiella and Pseudomonas resistance to Ampicillin was 85%, 91% and 100% respectively. The addition of the β-lactamase inhibitors (clavulanic acid or tazobactam) increases ampicillin activity (Co-amoxiclav such as Augmentin). This however was found to be less effective as it could only reduce the resistance in E.coli from 85 to 65%, in Klebsiella from 91 to 70% and in pseudomonas from 100 to 89%. The co-amoxiclave combination normally is very effective and increases the efficacy of ampicillin/amoxicillin to a quite valuable percentage. In a recent study in Riyadh 24, it was reported that the addition of β-lactamase inhibitor (clavulanic acid) restored the ampicillin activity (amoxicillin/clavulanate) in almost 37% of Gram negative bacteria. In the current study however, the maximum restoration of activity of the antibiotic was 21%.
Among Escherichia coli, resistance to Aztreonam, Imipenem, and Ciprofloxacin/Norfloxacin was significantly higher during the year 2017. The resistance to augmentin and cephalothin however decreased significantly during 2017. Being resistant to Aminoglycoside 6’-N-Acetyltransferase inactivation, the Amikacin was much more effective than gentamicin in E. coli (11% vs. 25%), Klebsiella (19% vs. 33%), and Pseudomonas (23% vs. 50%). Some other studies have also reported similar results22, 24
Klebsiella pneumoniae is one of the multidrug drug resistant bacteria considered as a serious threat to human health by the WHO and CDC. Infections caused by klebsiella pose a serious threat particularly among children, elderly and immunocompromised patients25, 26. Among the isolated Klebsiella spp. higher resistance levels were seen against most of the antibiotics. As described earlier, imipenem was the most effective antibiotic against klebsiella while Aztreonam faced significantly increased resistance during 2017.
Most common infections caused by enterococci are the UTI infections27. Higher resistance levels were seen among enterococci against most of the antibiotics. Surprisingly, augmentin and gentamicin were significantly more effective against enterococci during 2017.
Staphylococcal infections, particularly those caused by methicillin resistant S. aureus (MRSA), are increasing worldwide28-33. In the United States, MRSA is the most common cause of skin and soft-tissue infections34, 35. During the current study period, high levels of resistance among staphylococcus spp. were seen against most of the common antibiotic with significant increase in the resistance against Ampicillin, oxacillin, Cephalothin, cefotaxime, gentamicin and vancomycin during 2017.
Pseudomonas spp. frequently causes respiratory and urinary tract infections. Increasing resistance to cephalosporins, fluoroquinolones, carbapenems and other antibiotics have been reported in many studies16. In these studies, reported imipenem resistance was around 20% while in the current study it was found to be much less (<7%). The reported ciprofloxacin resistance (<“30%) however was less than that found in the current study (46%). The resistance against Piperacillin, cefuroxime, ceftazidime and ciprofloxacin/Norfloxacin was found to be significantly increasing during 2017.
According to the US Centers for Disease Control and Prevention (CDC), P. aeruginosa is the third most common cause of UTI. Almost 7% UTI infections are caused by this organism3
National Nosocomial Infections Surveillance (NNIS) system report: data summary from January 1992 through June 2003, issued August 200336.
In Saudi Arabia, carbapenem resistant Acinetobacter baumannii (CRAB) have also increased vividly in the recent years. Al-Obeid et al., 201537 revealed A. baumannii resistance to meropenem and imipenem in 2006 was between 20-35%, which increased to almost 90% during 2012. In the current study, A. baumannii resistance to imipenem was found in 8 out of 19 isolates (42%) during 2016. Molecular studies done on CRAB isolates from the GCC region showed that pathogenic isolates from different parts of the region have assembled together38.
Increased overall antibiotic resistance was observed in the current study. E. coli is the most prevalent uropathogen found in the study and this must be considered while selecting antimicrobial treatment for the UTIs. The resistance in the region is increasing against β-Lactam group except Augmentin where it showed opposite trend. Imipenem, Amikacin and Vancomycin are overall the most effective antibiotics against the uropathogens in the city.
As the antibiotic resistance is increasing by the time, routine urine cultures must be done for deciding a proper treatment for the infection to avoid treatment failure. Also, the antibiotic resistance trends must regularly be monitored in the region under a well-defined surveillance programs.
ACKNOWLEDGMENTS
The author gratefully acknowledges the approval and the support of this research study by the grant no 5791-PHM-2016-1-6-F from the Deanship of Scientific Research at Northern Border University, Arar, KSA.
- “About Antimicrobial Resistance – Antibiotic/Antimicrobial Resistance – CDC”. www.cdc.gov. 19 September 2017.
- Akram, M., Shahid, M., Khan, A.U., Etiology and antibiotic resistance patterns of community-acquired urinary tract infections in J N M C Hospital Aligarh, India. Ann Clin Microbiol Antimicrob., 2007; 6:4
- Williams, D.N., Urinary tract infection: Emerging insights into appropriate management. Postgrad Med, 1996; 99:189-92
- Stamm, W.E., Norrby, S.R. Urinary tract infections: disease panorama and challenges. J Infect Dis., 2001; 183(Suppl. 1), pp. S1-S4.
- Magalit, S.L., Gler, M.T., Tupasi, T.E., Increasing antimicrobial resistance patterns of community and nosocomial uropathogens in Makati Medical Center. Phil J Microbiol Infect Dis., 2004; 33: 143-8.
- Levinson, W., Jawetz, E., Medical microbiology and immunology: examination and board review (6th ed.), McGraw-Hill, New York.
- Valiquette, L., Urinary tract infections in women. Can J Urol., 2001; 1: pp. 6-12.
- Manges, A.R., Tabor, H., Tellis, P., Vincent, C., and Tellier, P., Endemic and Epidemic Lineages of Escherichia coli that Cause Urinary Tract Infections. Emerg Infect Dis., 2008; 14; 1575-1583.
- Farajnia, S., Alikhani, M,Y., Ghotaslou, R., Naghili, B., Nakhlband, A., Causative agents and antimicrobial susceptibilities of urinary tract infections in the northwest of Iran. Int J Infect Dis., 2009; 13: 140-4.
- Akortha, E.E., Ibadin, O.K., Incidence and antibiotic susceptibility pattern of Staphylococcus aureusamongst patients with urinary tract infection (UTI) in UBTH Benin City, Nigeria. Afr J Biotechnol., 2008; 7:1637-40.
- Zervos, M.J., Hershberger, E., Nicolau, D.P., Ritchie, D.J., Blackner, L.K., Coyle, E.A., et al., Relationship between fluoroquinolone use and changes in susceptibility to fluoroquinolones of selected pathogens in 10 United States teaching hospitals, 1991-2000. Clin Infect Dis., 2003; 37:1643–8
- Arslan, H., Azap, O.K., Ergönül, O., Timurkaynak, F., Risk factors for ciprofloxacin resistance among Escherichia colistrains isolated from community-acquired urinary tract infections in Turkey. J Antimicrob Chemother., 2005; 56: 914–8.
- Kurtaran, B., Candevir, A., Tasova, Y., Kibar, F., Inal, A. S., Komur S, Aksu HSZ. Antibiotic resistance in community-acquired urinary tract infections: Prevalence and risk factors. Med Sci Monit., 2010; 16(5): 246-251
- Khawcharoenporn, T., Vasoo, S., Singh, K., Urinary Tract Infections due to Multidrug-Resistant Enterobacteriaceae: Prevalence and Risk Factors in a Chicago. Emergency Medicine International, 2013; Article ID 258517, 7 pages,
- Sharma, I., and Paul, D., Prevalence of community acquired urinary tract infections (UTI) in Silchar Medical College, Assam, India and its antimicrobial susceptibility profile. Int. J. Microbiol. Res. Rev., 2013; 1 (1): 001-005.
- World Health Organization. Antimicrobial resistance: global report on surveillance. Geneva: WHO 2014; http://www.who.int/drugresistance/documents/surveillancereport/en/
- Lim, C.J., Cheng, A.C., Kong, D.C., Peleg, A.Y., Community onset bloodstream infection with multidrug resistant organisms: a matched case control study. BMC Infect Dis., 2014; 14: 126
- Stefaniuk, E., Suchocka, U., Bosacka, K., Hryniewicz, W., Etiology and antibiotic susceptibility of bacterial pathogens responsible for community acquired urinary tract infections in Poland. Eur J Clin Microbiol Infect Dis., 2016; 35: 1363–1369
- Lambert, M.L., Suetens, C., Savey, A., Palomar, M., Hiesmayr, M., Morales, I., Agodi, A., Frank, U., Mertens, K., Schumacher, M. et al., Clinical outcomes of health care associated infections and antimicrobial resistance in patients admitted to European intensive care units: a cohort study. Lancet Infect Dis., 2011; 11: 30–38
- Neidell, M.J., Cohen, B., Furuya, Y., Hill, J., Jeon, C.Y., Glied, S., Larson, E.L., Costs of healthcare and community associated infections with antimicrobial resistant versus antimicrobial susceptible organisms. Clin Infect Dis., 2012; 55: 807–815
- Martin Loeches, I., Torres, A., Rinaudo, M., Terraneo, S., de Rosa, F., Ramirez, P., Diaz, E., Fernandez Barat, L., Li Bassi, G.L., Ferrer, M., . Resistance patterns and outcomes in intensive care unit (ICU) acquired pneumonia. Validation of European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC) classification of multidrug resistant organisms. J Infect, 2015; 70: 213–222
- El-Kersh, T.A., Marie, M.A., Al-Sheikh, Y.A., and Al-Kahtani, S.A., Prevalence and risk factors of community-acquired urinary tract infections due to ESBL-producing Gram negative bacteria in an Armed Forces Hospital in Sothern Saudi Arabia. GARJMMS, 2015; 4(8): 321-330.
- Gupta, K., Hooton, T.M., Naber, K.G., Wullt, B., Colgan, R., Miller, L.G., Moran, G.J., Nicolle, L.E., Raz, R., Schaeffer, A.J., International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis., 2011; 52: e103–e120.
- Khadri, H., Alzohairy, M., High prevalence of multi-drugresistance (MDR) and extended spectrum b-lactamases (ESBL) producing bacteria among community-acquired urinary tract infections (CAUTI). J. Bacteriol. Res. 2009; 1(9): 105-110.
- Paczosa, M.K., Mecsas, J., Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev, 2016; 80: 629–661
- Quan, T.P., Fawcett, N.J., Wrightson, J.M., Finney, J., Wyllie, D., Jeffery, K., Jones, N., Shine, B., Clarke, L., Crook, D., et al., Increasing burden of community acquired pneumonia leading to hospitalisation, 1998–2014. Thorax, 2016; 71: 535–542
- Chakraborty, A., Pal, N.K., Sarkar, S., Gupta, M.S., Antibiotic resistance pattern of Enterococciisolates from nosocomial infections in a tertiary care hospital in Eastern India. Journal of Natural Science, Biology, and Medicine., 2015; 6(2): 394-397.
- Kaplan, S.L., et al., Three-year surveillance of community-acquired Staphylococcus aureus infections in children. Clin Infect Dis., 2005; 40: 1785–91.
- Hersh, A.L., Chambers, H.F., Maselli, J.H., Gonzales, R., National trends in ambulatory visits and antibiotic prescribing for skin and soft-tissue infections. Arch Intern Med., 2008; 168: 1585–91.
- Klevens, R.M., et al., Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama., 2007; 298:1763–71.
- Hope, R., Livermore, D.M., Brick, G., Lillie, M., Reynolds, R., Non-susceptibility trends among staphylococci from bacteraemias in the UK and Ireland, 2001–06. J Antimicrob Chemother., 2008; 62(2): ii65–74.
- Laupland, K.B., Ross, T., Gregson, D.B., Staphylococcus aureus bloodstream infections: risk factors, outcomes, and the influence of methicillin resistance in Calgary, Canada, 2000–2006. J Infect Dis., 2008; 198: 336–43.
- EARSS Annual Report. 2007. http://www.rivm.nl/earss/news/index.jsp.
- Moran, G.J., et al., (2006). Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med., 355:666–74.
- Fridkin, S,K., et al., Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med., 2005; 352:1436–44.
- http://www.cdc.gov/ncidod/dhqp/pdf (Retrieved on 15 January 2009)
- Al-Obeid, S., Jabri, L., Al-Agamy, M., Al-Omari, A., Shibl, A., Epidemiology of extensive drug resistant Acinetobacter baumannii (XDRAB) at Security Forces Hospital (SFH) in Kingdom of Saudi Arabia (KSA). J Chemother, 2015; 27: 156-162.
- Zowawi, H.M., Sartor, A.L., Sidjabat, H.E., Balkhy, H.H., Walsh, T.R., Al Johani, S.M., et al., Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf Cooperation Council States: dominance of OXA-23-type producers. J Clin Microbiol, 2015; 53: 896-903.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


