ISSN: 0973-7510
E-ISSN: 2581-690X
Microorganisms have broad spectrum of applications varying from metabolite production to breakdown of complex carbon sources. Cresols are one such complex that has drawn attention due to its disposal in un-degraded or partially degraded form that enters into air, water and soil polluting the environment; its extremities extend to surrounding ecosystem and destroying the aquatic and human life forms. Cresol possesses special affection towards blood plasma leading to kidney, liver and heart disorders. Many have reported biological degradation of hazardous chemical using different organisms with fine degradation efficiency. The current investigation adds another strain Pseudomonas monteilii which was found to exhibit the potential to utilize cresol as its major carbon source for growth and proliferation. Further, the Cresol inhibitory concentration was checked by varying concentration from 100 to 1500 ppm. The remnant concentration over the time period was analyzed by aminoantipyrine assay. The strain was capable of >99% removal at neutral pH, 150 rpm and 30p C in 24h. The phylogeny of the strain was analyzed post 16S rRNA sequencing using in silico tools. The strain can be optimized for degrading higher cresol dose without compromising on the biodegradation efficiency.
Cresol degrading bacteria, 16S rRNA Gene Sequencing, Pseudomonas monteilii.
The methylphenols are the aromatic organic compounds that have variable melting points decided by the surrounding temperatures. These methylphenols have various applications in manufacturing of pesticides, petroleum products, dyes and also in a few pharmaceutical products and are generally known as cresols1,2,3,4,5. Cresols are generally found dumped in regions with petroleum or dye dumping sites. They are severely toxic for humans if inhaled or ingested, even at a very low concentration that often leads to consequences varying from irritation of eyes, mouth, throat and skin, vomiting, liver and heart damage, paralysis, coma and death1,6,7. In human, cresol finds more affinity towards the ligand binding protein, Human Serum Albumin (HSA). HSA contributes as carrier proteins for various steroids, fatty acids and helps in maintaining the antioxidants in the body. Cresol binds to albumin reducing the carrier profile of protein resulting in kidney and blood disorders 8,9,10,11. It is not only toxic for human life but equally deleterious for the aquatic forms, thus, direct disposal in water, air and soil is strictly prohibited12,13,14.
The environment exposure to cresols has been observed in terms of petroleum leakage, sludge from dye, pharmaceutical or other industries using derivatives of cresols as raw material1. There have been a lot of investigation on degradation and sequestration of the hazardous compound, where the microorganisms including, Pseudomonas15, Acinetobacter16, Ralstonia17 etc. have been predominantly isolated from the contaminated sites. These microorganisms possess a strong specific mechanism to metabolize these phenols to a usable carbon source for their growth and proliferation. The enzymes such as monooxygenases, hydroxylases and dehydrogenases play a major role in formation of cresol intermediates and further, direct to Tri-carboxylic acid cycle (TCA). The end product thus, remains carbon dioxide and ATP, energy providing molecules18,19.
In the current study, a novel strain of Pseudomonas has been isolated form petroleum contaminated site, which has been found quite suitable for cresol degradation. The isolate has thus, been phylogentically analyzed.
Isolation and Screening of Cresol Degrading Bacteria
The soil samples were collected from the various localities in and around Calicut which were contaminated with petroleum, including, petrol pumps, automobile workshops, Petroleum transfer areas to the reservoir, etc. The 30 different soil types were inoculated in to Minimal mineral medium without carbon source as a negative control, whereas, for the test samples, various isomers of cresol were fed as carbon source in minimal mineral medium. The concentration of cresol isomers were maintained as low as 100 ppm in 1L of the medium. 1% of each soil sample was inoculated into the control and test samples and was incubated for24h at 30p C with 150 rpm. Depending on the microbial growth the concentration of cresol variants was gradually increased to 500 ppm for the consortium showing a good affinity towards the cresol minimal medium. 20
The cresol acclimatization by the grown consortium was tested at higher concentration ranging from 500 ppm to 1200 ppm. Based on the responses of each bacterial culture, the strain with best cresol tolerance and efficiency of survival on the cresol medium was isolated for phenotypic characterization.21
Morphological and Phenotypic Characterization
The isolated strain was undergone a series of biochemical and morphological identification. The strain was Gram stained and observed under the microscope to understand morphological characteristics. Further, the pigment producing efficiency, colony shape, aerobic requirements and other sugar utilizing efficiencies were tested.22
The study of phenotypical characterization was initiated with DNA isolation. The obtained sample was analyzed for purity and integrity on 2% agarose gel prepared in 1X TE buffer. PCR was run to amplify the product. Amplified product was sequenced using 16S rRNA sequencing.23 Phenotypical characterization was done in the Department of Life Sciences, Kristu Jayanti College, Bangalore. Phylogenetic analysis was performed using clustalW.
Cresol Degradation Study
The cresol degradation was checked by evaluating the remnant cresol concentration in the medium after the optimum incubation period. The evaluation of cresol content was performed by a colorimetric assay using 4-amino antipyrine method.24
Cresol was treated with 4-aminoantipyrine at alkaline pH around 10.00 in presence of an oxidizing agent, potassium ferricyanide that results in the formation of stable reddish-brown colored antipyrine dye. The dye can be estimated by taking O.D. at 460 nm. The intensity of color or the optical density is directly proportional to the concentration of cresol. The standard graph for the assay was plotted with known concentrations of cresol between 10 to 50µg/L. So, the O.D. values obtained for known concentration of cresol were extrapolated or interpolated on the standard curve to the quantitative cresol concentration.25,26,27
Isolation And Screening Of Cresol Degrading Bacteria
The thirty different soil types were inoculated in to minimal medium with and without cresol variants to grow the bacterial consortium possessed by each. The grown culture was found to have a good population of bacteria that was screened further based on the tolerance at high cresol concentration. Among the cresol variants used, o-Cresol was found to have better results in terms of microbial affinity. o-Cresol concentration was increased from 100ppm to 500ppm that resulted into survival of only 15 bacteria, namely CR-01 to CR-15. The o-Cresol tolerance was checked by varying concentration from 500ppm to 1200ppm. Most of the bacteria were found dead after 700ppm, with CR-13 showing exceedingly high tolerance for o-Cresol that survived even at 1200ppm. A similar study has been performed by Fatiha Lassouane et. al. on Pseudomonas aeruginosa strain (strain S8) where, the strain has a high o-Cresol biodegradation potential, it could degrade completely 1250 mg/ L of o-Cresol within 85 h.28
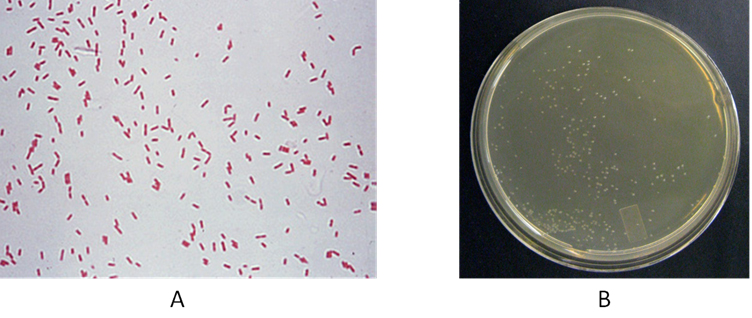
Fig. 1. (A) and (B)Gram staining of CR-13 and (B) wet, circular and smooth colonies of CR-13 on minimal mineral medium supplemented with 500 ppm of o-cresol.
Thereafter, CR-13 was characterized with various microscopic features, (Table 1) colony characters (Table 2) and biochemical tests (Table 3). The strain was found to be Gram negative, rod-shaped motile organism, without spore and capsule. [Figure 1 (A)]. It possessed circular colonies with smooth and wet texture, [Figure 1 (B)]. This aerobic bacterium was found catalase and oxidase positive.
Table (1):
Microscopic features of CR-13.
Sl. No. |
Features |
Isolates |
|---|---|---|
1 |
Gram’s staining |
Gram positive |
2 |
Motility |
Motile |
3 |
Endospore |
present |
4 |
Capsule |
Absent |
Table (2):
Colony characteristics of CR-13.
| Sl. No. | Colony morphology | |
|---|---|---|
| 1 | Size | Pin point |
| 2 | Shape | Circular |
| 3 | Margin | Smooth, Entire |
| 4 | Elevation | Raised and Convex |
| 5 | Optical features | Wet and Translucent |
| 6 | Pigmentation | NIL |
Table (3):
Biochemical characterization of CR-13.
Sl. No. |
Biochemical tests |
Result |
|---|---|---|
1 |
Indole production test |
Negative (-ve) |
2 |
Methyl Red |
Negative (-ve) |
3 |
Voges Proskauer test |
Negative (-ve) |
4 |
Citrate Utilization |
Positive (+ve) |
5 |
Oxygen requirement |
Aerobic |
6 |
catalase |
Positive (+ve) |
7 |
oxidase |
Positive (+ve) |
8 |
Dextrose Fermentation |
Positive (+ve) |
9 |
Sucrose Fermentation |
Positive (+ve) |
10 |
Lactose Fermentation |
Negative (-ve) |
Phylogenetic Characterization
The sequence, received from the sequencer was converted into FASTA file using bioinformatics software. The sequence then used for Blast search against NCBI and phylogenetic analysis was performed using clustalW. The phylogenetic analysis predicted that the sequence, obtained was found to have 99% similarity with Pseudomonas monteilii strain MBG2 16S ribosomal RNA gene. (Figure 2).
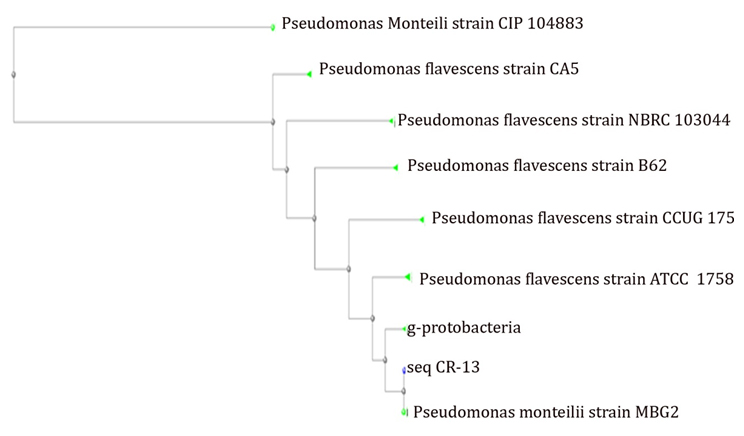
Fig. 2. Phylogenetic analysis of seq. CR-13 using CLUSTAL-W
The growth profile of Pseudomonas monteilii CR-13 at different concentration of o-Cresol was studied in the presence and absence of standard carbon source, cresol and without any carbon source. The growth of the cells was studied using UV-Vis spectrophotometer at 600nm. (Figure 3). All the varied initial concentrations of o-Cresol were consumed by P. monteilii CR-13. The P. monteilii CR-13 could survive and utilize upto 1500ppm of o-Cresol with a low growth profile than the standard carbon source (glucose). Similar studies have reported by Ahamad, P.Y.A. et al.29,and Mamma D30 where acclimatized Pseudomonas putida cells could overcome the inhibitory effect of phenol by the addition of glucose, a conventional carbon source.
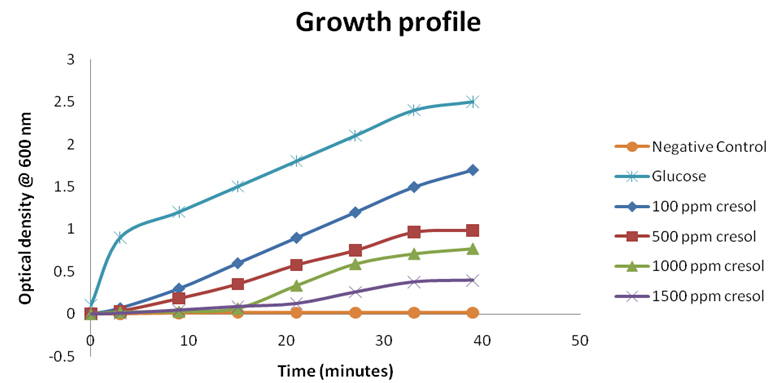
Fig. 3. Growth profile of Pseudomonas monteilii CR 13 at the different initial concentrations of o-cresol
Degradation Efficiency of P. monteilii CR13
The biodegradation study of cresol by P. monteilii CR13 was performed by 4-amino-antipyrine assay. The biodegradation study in terms of residual concentration of o-Cresol was performed and the unknown concentration was calculated based on the optical density of the standard curve for 4-aminoantipyrine assay. The maximum degradation was affected by the initial cresol concentration in the medium. The amount of remnant cresol concentration was determined by interpolating the O.D. values obtained after 4-amino antipyrine assay. The values of O.D. at different time intervals between 0 to 30 h have been plotted over the standard curve to determine the unknown concentrations. So, by the end of each experimental set, the concentrations calculated was 0.25, 2.5, 2.75 and 5 ppm, respectively with initial cresol dose of 100, 500, 1000 and 1500 ppm. The degradation efficiencies achieved are 99.75, 99.5, 99.35 and 98.28, respectively for 100 ppm, 500 ppm , 1000 ppm, and 1500 ppm respectively. (Figure 4)
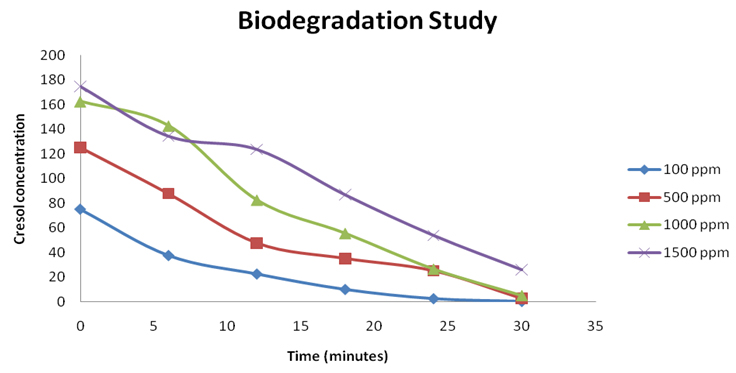
Fig. 4. Initial concentration of cresol influences the degradation efficiency of pseudomonas monteilii CR-13
The isolated Pseudomonas monteilii CR-13 is an efficient bacterial strain to biodegrade o-Cresol. The P. monteilii CR-13 is efficiently degrading 1500 ppm of o-Cresol and shows good growth profile. Further investigations are recommended for immobilization and optimizing the growth conditions for enhancing the degrading capacity of P. monteilii CR-13. In the light of results that were gathered from our batch culture, the organism is showing high promise in degradation of o-Cresol.
- Al-Khalid, T., El-Naas, M. H. Aerobic biodegradation of phenols: a comprehensive review Critical Reviews in Environmental Science and Technology, 2012; 42(16): 1631-1690.
- Chaurasia, A. K., Tremblay, P., Holmes, D. E., Zhang, T. Genetic evidence that the degradation of para-cresol by Geobacter metallireducens is catalyzed by the periplasmic para-cresol methylhydroxylase. FEMS microbiology letters, 2015; 362(20): fnv145.
- Wang, Y., Wei, H., Zhao, Y., Sun, W., Sun, C. The optimization, kinetics and mechanism of m-cresol degradation via catalytic wet peroxide oxidation with sludge-derived carbon catalyst. Journal of Hazardous Materials, 2017; 326: 36-46. DOI: PMID:27987448
- Balachandran, R., Patterson, Z., Deymier, P., Snyder, S. A., Keswani, M. Understanding acoustic cavitation for sonolytic degradation of p-cresol as a model contaminant. Chemosphere, 2016; 147:52-59
- Fiege, H., Voges, H. W., Hamamoto, T., Umemura, S., Iwata, T., Miki, H., Fujita, Y., Buysch, H. J., Garbe, D., Paulus, W. Phenol derivatives. Ullmann’s Encyclopedia of Industrial Chemistry, 2000.
- Nair,C. I., Jayachandran, K., Shashidhar, S. Biodegradation of phenol. African Journal of Biotechnology. 2008; 7(25): 4951-4958
- Zhao, G., Zhou, L., Li, Y., Liu, X., Ren, X. Enhancement of phenol degradation using immobilized microorganisms and organic modified montmorillonite in a two-phase partitioning bioreactor.J. Hazardous Mater. 2009; 169:402-410. DOI: 10.1016/j.jhazmat.2009.03.110
- Meijers, B., Bammens, B., Moor, B., De, Verbeke, K., Vanrenterghem, Y. Eveepoel, P. Free p-cresol is associated with cardiovascular disease in hemodialysis patients.Kidney international, 2008; 73(10): 1174-1180.
- Meijers, B. K., Claes, K., Bammens, B., Loor, H. De, Viaene, L., Verbeke, K., Vanrenterghem, Y., Evenepoel, P. p-Cresol and cardiovascular risk in mild-to-moderate kidney disease. Clinical journal of the American Society of Nephrology, 2010; 5(7): 1182-1189.
- Maeda, M., Itoh, A., Kawase, Y. Kinetics for aerobic biological treatment of o-cresol containing wastewaters in a slurry bioreactor: biodegradation by utilizing waste activated sludge. Biochemical Engineering Journal, 2005; 22(2): 97-103.
- Agarwal, R., Davis, J. L., Smith, L. Serum albumin is strongly associated with erythropoietin sensitivity in hemodialysis patients. Clinical Journal of the American Society of Nephrology, 2008; 3(1): 98-104.
- Braunbeck, T. and Völkl, A., Toxicant-induced cytological alterations in fish liver as biomarkers of environmental pollution? A case study on hepatocellular effects of dinitro-o-cresol in golden ide (Leuciscus idus melanotus). Fish-ecotoxicology and ecophysiology, 1993, 55-80.
- Versino, B., De Groot, M., Geiss, F. Air pollution—sampling by adsorption columns. Chromatographia. 1974; 7(6): 302-304.
- Evangelista, R. A., Allen, H. L., Manuel, R. M. Treatment of phenol and cresol contaminated soil. Journal of hazardous materials, 1990; 25(3): 343-360.
- Ahamad, P., Kunhi, A.A.M., Divakar, S. New metabolic pathway for o-cresol degradation by Pseudomonas sp. CP4 as evidenced by 1H NMR spectroscopic studies. World Journal of Microbiology and Biotechnology; 2001; 17(4): 371-377.
- Liu, Y., Zhang, A., Wang, X. Biodegradation of phenol by using free and immobilized cells of Acinetobacter sp. XA05 and Sphingomonas sp. FG03. Biochemical Engineering Journal, 2009; 44(2): 187-192.
- Bai, J., Wen, J. P., Li, H. M., Jiang,Y. Kinetic modeling of growth and biodegradation of phenol and m-cresol using Alcaligenes faecalis, Process Biochem. 2007; 42:10–517.
- Hopper, D., Taylor, D. Pathways for the degradation of m-cresol and p-cresol by Pseudomonas putida. Journal of bacteriology. 1975;1-6.
- Wei, X., Tetyana, G., Felix, W., Richnow, H., Carsten, V. Characterization of phenol and cresol biodegradation by compound-specific stable isotope analysis. Environmental Pollution. 2016; 166-173.
- Hamitouche, A., Bendjama, Z., Amrane, A., Kaouah, F. Biodegradation of p-cresol by Pseudomonas spp. Desalination and Water Treatment. 2016; 57(7): 3059-3064.
- Spain, J. C., Van Veld, P. Adaptation of natural microbial communities to degradation of xenobiotic compounds: effects of concentration, exposure time, inoculum, and chemical structure. Applied and Environmental Microbiology. Applied and Environmental Microbiology, 1983; 45(2): 428-435.
- Daane, L. L., Harjono, I., Zylstra, G. J., Haggblom, M. M. Isolation and characterization of polycyclic aromatic hydrocarbon-degrading bacteria associated with the rhizosphere of salt marsh plants. Applied and Environmental Microbiology, 2001; 67(6): 2683-2691
- Heinaru, E., Naanuri, E., Grunbach, M., Joesaar, M., Heinaru, A. Functional redundancy in phenol and toluene degradation in Pseudomonas stutzeri strains isolated from the Baltic Sea. Gene, 2016, 589(1), 90-98.
- Hannaford, A. M., Kuek, C. Aerobic batch degradation of phenol using immobilized Pseudomonas putida. Journal of industrial microbiology & biotechnology, 1999; 22(2): 121-126.
- Kind, P., King, E. Estimation of plasma phosphatase by determination of hydrolysed phenol with amino-antipyrine. Journal of clinical Pathology, 1954; 7(4): 322.
- Ettinger. M., Ruchhoft, C., Lishka, R. Sensitive 4-aminoantipyrine method for phenolic compounds. Analytical Chemistry, 1951; 23(12):1783-1788.
- Abbassian. K., Kargari, A., Kaghazchi, T. Phenol removal from aqueous solutions by a novel industrial solvent. Chemical Engineering Communications, 2015; 202: 408-413.
- Lassouane, F., Amrani, S., Aït-Amar, H. Evaluation of o-cresol degradation potential by a strainof Pseudomonas aeruginosa S8. Desalination And Water Treatment, 2013; 51(40-42): 7577-7585.
- Ahamad. P. Y. A., Kunhi, A. A. M. Degradation of high concentrations of cresols byPseudomonas sp. CP4. World Journal of Microbiology and Biotechnology, 1999; 15: 281–283
- Mamma, D., Kalogeris, E., Papadopoulos, N., Hatzinikolaou, D. G., Christrakopoulos, P., Kekos, D. Biodegradation of phenol by acclimatized Pseudomonas putida cells using glucose as an added growth substrate. Journal of Environmental Science and Health, 2004; 39(8): 2093-2104.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


